- Record: found
- Abstract: found
- Article: found
Purinergic Receptors in the Airways: Potential Therapeutic Targets for Asthma?
Read this article at
Abstract
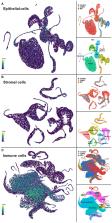
Extracellular ATP functions as a signaling messenger through its actions on purinergic receptors, and is known to be involved in numerous physiological and pathophysiological processes throughout the body, including in the lungs and airways. Consequently, purinergic receptors are considered to be promising therapeutic targets for many respiratory diseases, including asthma. This review explores how online bioinformatics resources combined with recently generated datasets can be utilized to investigate purinergic receptor gene expression in tissues and cell types of interest in respiratory disease to identify potential therapeutic targets, which can then be investigated further. These approaches show that different purinergic receptors are expressed at different levels in lung tissue, and that purinergic receptors tend to be expressed at higher levels in immune cells and at more moderate levels in airway structural cells. Notably, P2RX1, P2RX4, P2RX7, P2RY1, P2RY11, and P2RY14 were revealed as the most highly expressed purinergic receptors in lung tissue, therefore suggesting that these receptors have good potential as therapeutic targets for asthma and other respiratory diseases.
Related collections
Most cited references81
- Record: found
- Abstract: found
- Article: not found
Proteomics. Tissue-based map of the human proteome.
- Record: found
- Abstract: found
- Article: not found
Gene Expression Omnibus: NCBI gene expression and hybridization array data repository.
- Record: found
- Abstract: found
- Article: not found