- Record: found
- Abstract: found
- Article: not found
Mitochondrial Dynamics in SARS-COV2 Spike Protein Treated Human Microglia: Implications for Neuro-COVID

Read this article at
Abstract
Abstract
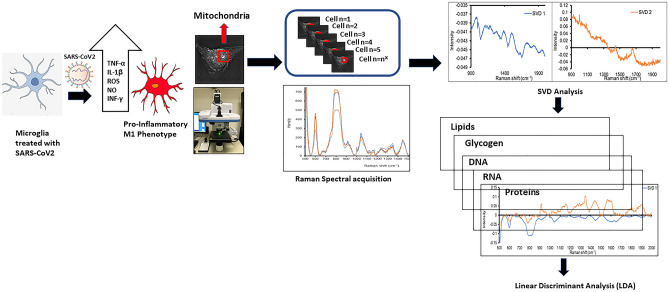
Emerging clinical data from the current COVID-19 pandemic suggests that ~ 40% of COVID-19 patients develop neurological symptoms attributed to viral encephalitis while in COVID long haulers chronic neuro-inflammation and neuronal damage result in a syndrome described as Neuro-COVID. We hypothesize that SAR-COV2 induces mitochondrial dysfunction and activation of the mitochondrial-dependent intrinsic apoptotic pathway, resulting in microglial and neuronal apoptosis. The goal of our study was to determine the effect of SARS-COV2 on mitochondrial biogenesis and to monitor cell apoptosis in human microglia non-invasively in real time using Raman spectroscopy, providing a unique spatio-temporal information on mitochondrial function in live cells. We treated human microglia with SARS-COV2 spike protein and examined the levels of cytokines and reactive oxygen species (ROS) production, determined the effect of SARS-COV2 on mitochondrial biogenesis and examined the changes in molecular composition of phospholipids. Our results show that SARS- COV2 spike protein increases the levels of pro-inflammatory cytokines and ROS production, increases apoptosis and increases the oxygen consumption rate (OCR) in microglial cells. Increases in OCR are indicative of increased ROS production and oxidative stress suggesting that SARS-COV2 induced cell death. Raman spectroscopy yielded significant differences in phospholipids such as Phosphatidylinositol (PI), phosphatidylserine (PS), phosphatidylethanolamine (PE) and phosphatidylcholine (PC), which account for ~ 80% of mitochondrial membrane lipids between SARS-COV2 treated and untreated microglial cells. These data provide important mechanistic insights into SARS-COV2 induced mitochondrial dysfunction which underlies neuropathology associated with Neuro-COVID.
Related collections
Most cited references71
- Record: found
- Abstract: found
- Article: not found
Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method.
- Record: found
- Abstract: found
- Article: not found
SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor

- Record: found
- Abstract: found
- Article: found