- Record: found
- Abstract: found
- Article: found
miR-20b and miR-451a Are Involved in Gastric Carcinogenesis through the PI3K/AKT/mTOR Signaling Pathway: Data from Gastric Cancer Patients, Cell Lines and Ins-Gas Mouse Model
Read this article at
Abstract
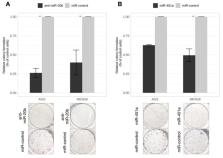
Gastric cancer (GC) is one of the most common and lethal gastrointestinal malignancies worldwide. Many studies have shown that development of GC and other malignancies is mainly driven by alterations of cellular signaling pathways. MicroRNAs (miRNAs) are small noncoding molecules that function as tumor-suppressors or oncogenes, playing an essential role in a variety of fundamental biological processes. In order to understand the functional relevance of miRNA dysregulation, studies analyzing their target genes are of major importance. Here, we chose to analyze two miRNAs, miR-20b and miR-451a, shown to be deregulated in many different malignancies, including GC. Deregulated expression of miR-20b and miR-451a was determined in GC cell lines and the INS-GAS mouse model. Using Western Blot and luciferase reporter assay we determined that miR-20b directly regulates expression of PTEN and TXNIP, and miR-451a: CAV1 and TSC1. Loss-of-function experiments revealed that down-regulation of miR-20b and up-regulation of miR-451a expression exhibits an anti-tumor effect in vitro (miR-20b: reduced viability, colony formation, increased apoptosis rate, and miR-451a: reduced colony forming ability). To summarize, the present study identified that expression of miR-20b and miR-451a are deregulated in vitro and in vivo and have a tumor suppressive role in GC through regulation of the PI3K/AKT/mTOR signaling pathway.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: not found
MicroRNAs in human cancer.
- Record: found
- Abstract: found
- Article: not found
Molecular evolution of a microRNA cluster.
- Record: found
- Abstract: found
- Article: not found
The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo.
Author and article information
Comments
Comment on this article
 Smart Citations
Smart CitationsSee how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.