- Record: found
- Abstract: found
- Article: found
Assessing Bacterial Populations in the Lung by Replicate Analysis of Samples from the Upper and Lower Respiratory Tracts
Read this article at
Abstract
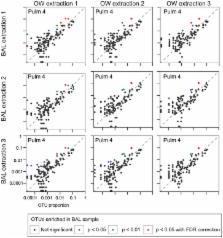
Microbes of the human respiratory tract are important in health and disease, but accurate sampling of the lung presents challenges. Lung microbes are commonly sampled by bronchoscopy, but to acquire samples the bronchoscope must pass through the upper respiratory tract, which is rich in microbes. Here we present methods to identify authentic lung microbiota in bronchoalveolar lavage (BAL) fluid that contains substantial oropharyngeal admixture. We studied clinical BAL samples from six selected subjects with potential heavy lung colonization. A single sample of BAL fluid was obtained from each subject along with contemporaneous oral wash (OW) to sample the oropharynx, and then DNA was extracted from three separate aliquots of each. Bacterial 16S rDNA sequences were amplified and products analyzed by 454 pyrosequencing. By comparing replicates, we were able to specify the depth of sequencing needed to reach a 95% chance of identifying a bacterial lineage of a given proportion—for example, at a depth of 5,000 tags, OTUs of proportion 0.3% or greater would be called with 95% confidence. We next constructed a single-sided outlier test that allowed lung-enriched organisms to be quantified against a background of oropharyngeal admixture, and assessed improvements available with replicate sequence analysis. This allowed identification of lineages enriched in lung in some BAL specimens. Finally, using samples from healthy volunteers collected at multiple sites in the upper respiratory tract, we show that OW provides a reasonable but not perfect surrogate for bacteria carried into to the lung by a bronchoscope. These methods allow identification of microbes that can replicate in the lung despite the background due to oropharyngeal microbes derived from aspiration and bronchoscopic carry-over.
Related collections
Most cited references25

- Record: found
- Abstract: found
- Article: found
Dirichlet Multinomial Mixtures: Generative Models for Microbial Metagenomics
- Record: found
- Abstract: found
- Article: not found
Accurate determination of microbial diversity from 454 pyrosequencing data.
- Record: found
- Abstract: found
- Article: not found