- Record: found
- Abstract: found
- Article: not found
Targeting neuronal activity-regulated neuroligin-3 dependency in high-grade glioma
Read this article at
Summary
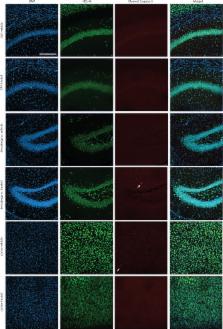
High-grade gliomas (HGG) are a devastating group of cancers, representing the leading cause of brain tumor-related death in both children and adults. Therapies aimed at mechanisms intrinsic to the glioma cell have translated to only limited success; effective therapeutic strategies will need to also target elements of the tumor microenvironment that promote glioma progression. We recently demonstrated that neuronal activity robustly promotes the growth of a range of molecularly and clinically distinct HGG types, including adult glioblastoma (GBM), anaplastic oligodendroglioma, pediatric GBM, and diffuse intrinsic pontine glioma (DIPG) 1 . An important mechanism mediating this neural regulation of brain cancer is activity-dependent cleavage and secretion of the synaptic molecule neuroligin-3 (NLGN3), which promotes glioma proliferation through the PI3K-mTOR pathway 1 . However, neuroligin-3 necessity to glioma growth, proteolytic mechanism of secretion and further molecular consequences in glioma remain to be clarified. Here, we demonstrate a striking dependence of HGG growth on microenvironmental neuroligin-3, elucidate signaling cascades downstream of neuroligin-3 binding in glioma and determine a therapeutically targetable mechanism of secretion. Patient-derived orthotopic xenografts of pediatric GBM, DIPG and adult GBM fail to grow in Nlgn3 knockout mice. Neuroligin-3 stimulates numerous oncogenic pathways, including early focal adhesion kinase activation upstream of PI3K-mTOR, and induces transcriptional changes including upregulation of numerous synapse-related genes in glioma cells. Neuroligin-3 is cleaved from both neurons and oligodendrocyte precursor cells via the ADAM10 sheddase. ADAM10 inhibitors prevent release of neuroligin-3 into the tumor microenvironment and robustly block HGG xenograft growth. This work defines a promising strategy for targeting neuroligin-3 secretion, which could prove transformative for HGG therapy.
Related collections
Most cited references29
- Record: found
- Abstract: found
- Article: found
featureCounts: An efficient general-purpose program for assigning sequence reads to genomic features
- Record: found
- Abstract: found
- Article: not found
A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice.
- Record: found
- Abstract: not found
- Article: not found
