- Record: found
- Abstract: found
- Article: found
A curated and standardized adverse drug event resource to accelerate drug safety research
Read this article at
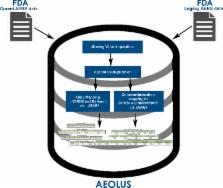
Abstract
Identification of adverse drug reactions (ADRs) during the post-marketing phase is one of the most important goals of drug safety surveillance. Spontaneous reporting systems (SRS) data, which are the mainstay of traditional drug safety surveillance, are used for hypothesis generation and to validate the newer approaches. The publicly available US Food and Drug Administration (FDA) Adverse Event Reporting System (FAERS) data requires substantial curation before they can be used appropriately, and applying different strategies for data cleaning and normalization can have material impact on analysis results. We provide a curated and standardized version of FAERS removing duplicate case records, applying standardized vocabularies with drug names mapped to RxNorm concepts and outcomes mapped to SNOMED-CT concepts, and pre-computed summary statistics about drug-outcome relationships for general consumption. This publicly available resource, along with the source code, will accelerate drug safety research by reducing the amount of time spent performing data management on the source FAERS reports, improving the quality of the underlying data, and enabling standardized analyses using common vocabularies.
Related collections
Most cited references29
- Record: found
- Abstract: found
- Article: not found
Use of proportional reporting ratios (PRRs) for signal generation from spontaneous adverse drug reaction reports.
- Record: found
- Abstract: found
- Article: not found
Utilizing social media data for pharmacovigilance: A review.

- Record: found
- Abstract: found
- Article: found