- Record: found
- Abstract: found
- Article: found
Identification of candidate flavonoid pathway genes using transcriptome correlation network analysis in ripe strawberry ( Fragaria × ananassa) fruits
Read this article at
Highlight
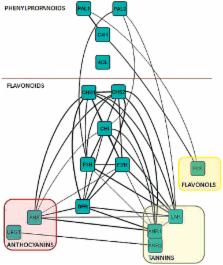
This work in strawberry fruit predicts new genes relevant to a key metabolic pathway using correlation analysis, and then demonstrates their function in a rapid and effective transient assay.
Abstract
New modulators of the strawberry flavonoid pathway were identified through correlation network analysis. The transcriptomes of red, ripe fruit from two parental lines and 14 of their progeny were compared, and uncharacterized transcripts matching the expression patterns of known flavonoid-pathway genes were identified. Fifteen transcripts corresponded with putative transcription factors, and several of these were examined experimentally using transient expression in developing fruits. The results suggest that two of the newly-identified regulators likely contribute to discrete nodes of the flavonoid pathway. One increases only LEUCOANTHOCYANIDIN REDUCTASE ( LAR) and FLAVONOL 3’-HYDROXYLASE (F3’H) transcript accumulation upon overexpression. Another affects LAR and FLAVONOL SYNTHASE (FLS) after overexpression. The third putative transcription factor appears to be a universal regulator of flavonoid-pathway genes, as many pathway transcripts decrease in abundance when this gene is silenced. This report demonstrates that such systems-level approaches may be especially powerful when connected to an effective transient expression system, helping to provide rapid and strong evidence of gene function in key fruit-ripening processes.
Related collections
Most cited references15
- Record: found
- Abstract: found
- Article: not found
Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits.
- Record: found
- Abstract: found
- Article: not found
The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes.
- Record: found
- Abstract: found
- Article: not found