- Record: found
- Abstract: found
- Article: found
Cradles and museums of generic plant diversity across tropical Africa
Read this article at
Summary
-
Determining where species diversify (cradles) and persist (museums) over evolutionary time is fundamental to understanding the distribution of biodiversity and for conservation prioritization. Here, we identify cradles and museums of angiosperm generic diversity across tropical Africa, one of the most biodiverse regions on Earth.
-
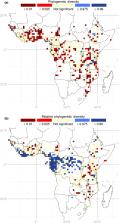
Regions containing nonrandom concentrations of young (neo‐) and old (paleo‐) endemic taxa were identified using distribution data of 1719 genera combined with a newly generated time‐calibrated mega‐phylogenetic tree. We then compared the identified regions with the current network of African protected areas (PAs).
-
At the generic level, phylogenetic diversity and endemism are mainly concentrated in the biogeographically complex region of Eastern Africa. We show that mountainous areas are centres of both neo‐ and paleo‐endemism. By contrast, the Guineo‐Congolian lowland rain forest region is characterized by widespread and old lineages. We found that the overlap between centres of phylogenetic endemism and PAs is high (> 85%).
-
We show the vital role played by mountains acting simultaneously as cradles and museums of tropical African plant biodiversity. By contrast, lowland rainforests act mainly as museums for generic diversity. Our study shows that incorporating large‐scale taxonomically verified distribution datasets and mega‐phylogenies lead to an improved understanding of tropical plant biodiversity evolution.
Related collections
Most cited references78
- Record: found
- Abstract: found
- Article: not found
Preserving the evolutionary potential of floras in biodiversity hotspots.
- Record: found
- Abstract: found
- Article: not found
The age and diversification of the angiosperms re-revisited.
- Record: found
- Abstract: found
- Article: not found