- Record: found
- Abstract: found
- Article: found
OptZyme: Computational Enzyme Redesign Using Transition State Analogues
Read this article at
Abstract
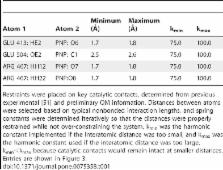
OptZyme is a new computational procedure for designing improved enzymatic activity (i.e., k cat or k cat/K M) with a novel substrate. The key concept is to use transition state analogue compounds, which are known for many reactions, as proxies for the typically unknown transition state structures. Mutations that minimize the interaction energy of the enzyme with its transition state analogue, rather than with its substrate, are identified that lower the transition state formation energy barrier. Using Escherichia coli β-glucuronidase as a benchmark system, we confirm that K M correlates (R 2 = 0.960) with the computed interaction energy between the enzyme and the para-nitrophenyl- β, D-glucuronide substrate, k cat/K M correlates (R 2 = 0.864) with the interaction energy of the transition state analogue, 1,5-glucarolactone, and k cat correlates (R 2 = 0.854) with a weighted combination of interaction energies with the substrate and transition state analogue. OptZyme is subsequently used to identify mutants with improved K M, k cat, and k cat/K M for a new substrate, para-nitrophenyl- β, D-galactoside. Differences between the three libraries reveal structural differences that underpin improving K M, k cat, or k cat/K M. Mutants predicted to enhance the activity for para-nitrophenyl- β, D-galactoside directly or indirectly create hydrogen bonds with the altered sugar ring conformation or its substituents, namely H162S, L361G, W549R, and N550S.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
Kemp elimination catalysts by computational enzyme design.
- Record: found
- Abstract: found
- Article: not found