- Record: found
- Abstract: found
- Article: found
Plasmodium falciparum hydroxymethylbilane synthase does not house any cosynthase activity within the haem biosynthetic pathway
Read this article at
Abstract
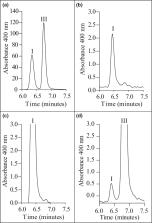
Uroporphyrinogen III, the universal progenitor of macrocyclic, modified tetrapyrroles, is produced from aminolaevulinic acid (ALA) by a conserved pathway involving three enzymes: porphobilinogen synthase (PBGS), hydroxymethylbilane synthase (HmbS) and uroporphyrinogen III synthase (UroS). The gene encoding uroporphyrinogen III synthase has not yet been identified in Plasmodium falciparum, but it has been suggested that this activity is housed inside a bifunctional hybroxymethylbilane synthase (HmbS). Additionally, an unknown protein encoded by PF3D7_1247600 has also been predicted to possess UroS activity. In this study it is demonstrated that neither of these proteins possess UroS activity and the real UroS remains to be identified. This was demonstrated by the failure of codon-optimized genes to complement a defined Escherichia coli hemD − mutant (SASZ31) deficient in UroS activity. Furthermore, HPLC analysis of the oxidized reaction product from recombinant, purified P. falciparum HmbS showed that only uroporphyrin I could be detected (corresponding to hydroxymethylbilane production). No uroporphyrin III was detected, showing that P. falciparum HmbS does not have UroS activity and can only catalyze the formation of hydroxymethylbilane from porphobilinogen.
Related collections
Most cited references27
- Record: found
- Abstract: found
- Article: not found
The biochemistry of heme biosynthesis.
- Record: found
- Abstract: found
- Article: not found
The heme biosynthesis pathway is essential for Plasmodium falciparum development in mosquito stage but not in blood stages.
- Record: found
- Abstract: found
- Article: not found