- Record: found
- Abstract: found
- Article: found
Deciphering the complexities of the wheat flour proteome using quantitative two-dimensional electrophoresis, three proteases and tandem mass spectrometry
Read this article at
Abstract
Background
Wheat flour is one of the world's major food ingredients, in part because of the unique end-use qualities conferred by the abundant glutamine- and proline-rich gluten proteins. Many wheat flour proteins also present dietary problems for consumers with celiac disease or wheat allergies. Despite the importance of these proteins it has been particularly challenging to use MS/MS to distinguish the many proteins in a flour sample and relate them to gene sequences.
Results
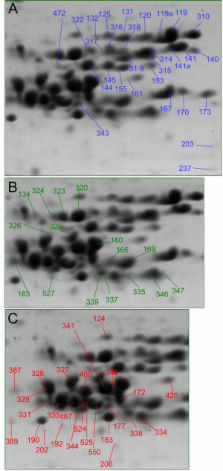
Grain from the extensively characterized spring wheat cultivar Triticum aestivum 'Butte 86' was milled to white flour from which proteins were extracted, then separated and quantified by 2-DE. Protein spots were identified by separate digestions with three proteases, followed by tandem mass spectrometry analysis of the peptides. The spectra were used to interrogate an improved protein sequence database and results were integrated using the Scaffold program. Inclusion of cultivar specific sequences in the database greatly improved the results, and 233 spots were identified, accounting for 93.1% of normalized spot volume. Identified proteins were assigned to 157 wheat sequences, many for proteins unique to wheat and nearly 40% from Butte 86. Alpha-gliadins accounted for 20.4% of flour protein, low molecular weight glutenin subunits 18.0%, high molecular weight glutenin subunits 17.1%, gamma-gliadins 12.2%, omega-gliadins 10.5%, amylase/protease inhibitors 4.1%, triticins 1.6%, serpins 1.6%, purinins 0.9%, farinins 0.8%, beta-amylase 0.5%, globulins 0.4%, other enzymes and factors 1.9%, and all other 3%.
Conclusions
This is the first successful effort to identify the majority of abundant flour proteins for a single wheat cultivar, relate them to individual gene sequences and estimate their relative levels. Many genes for wheat flour proteins are not expressed, so this study represents further progress in describing the expressed wheat genome. Use of cultivar-specific contigs helped to overcome the difficulties of matching peptides to gene sequences for members of highly similar, rapidly evolving storage protein families. Prospects for simplifying this process for routine analyses are discussed. The ability to measure expression levels for individual flour protein genes complements information gained from efforts to sequence the wheat genome and is essential for studies of effects of environment on gene expression.
Related collections
Most cited references70

- Record: found
- Abstract: found
- Article: found
Alpha-gliadin genes from the A, B, and D genomes of wheat contain different sets of celiac disease epitopes
- Record: found
- Abstract: found
- Article: not found