- Record: found
- Abstract: found
- Article: found
Genetic, morphometric, and molecular analyses of interspecies differences in head shape and hybrid developmental defects in the wasp genus Nasonia
Abstract
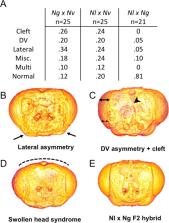
Males in the parasitoid wasp genus Nasonia have distinct, species-specific, head shapes. The availability of fertile hybrids among the species, along with obligate haploidy of males, facilitates analysis of complex gene interactions in development and evolution. Previous analyses showed that both the divergence in head shape between Nasonia vitripennis and Nasonia giraulti, and the head-specific developmental defects of F2 haploid hybrid males, are governed by multiple changes in networks of interacting genes. Here, we extend our understanding of the gene interactions that affect morphogenesis in male heads. Use of artificial diploid male hybrids shows that alleles mediating developmental defects are recessive, while there are diverse dominance relationships among other head shape traits. At the molecular level, the sex determination locus doublesex plays a major role in male head shape differences, but it is not the only important factor. Introgression of a giraulti region on chromsome 2 reveals a recessive locus that causes completely penetrant head clefting in both males and females in a vitripennis background. Finally, a third species ( N. longicornis) was used to investigate the timing of genetic changes related to head morphology, revealing that most changes causing defects arose after the divergence of N. vitripennis from the other species, but prior to the divergence of N. giraulti and N. longicornis from each other. Our results demonstrate that developmental gene networks can be dissected using interspecies crosses in Nasonia, and set the stage for future fine-scale genetic dissection of both head shape and hybrid developmental defects.
Related collections
Most cited references57
- Record: found
- Abstract: found
- Article: not found
Epistasis--the essential role of gene interactions in the structure and evolution of genetic systems.
- Record: found
- Abstract: not found
- Article: not found
Extensions of the Procrustes Method for the Optimal Superimposition of Landmarks
- Record: found
- Abstract: found
- Article: not found