- Record: found
- Abstract: found
- Article: found
Genetic variations in ARE1 mediate grain yield by modulating nitrogen utilization in rice
Read this article at
Abstract
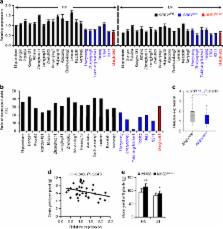
In crops, nitrogen directly determines productivity and biomass. However, the improvement of nitrogen utilization efficiency (NUE) is still a major challenge in modern agriculture. Here, we report the characterization of are1, a genetic suppressor of a rice fd-gogat mutant defective in nitrogen assimilation. ARE1 is a highly conserved gene, encoding a chloroplast-localized protein. Loss-of-function mutations in ARE1 cause delayed senescence and result in 10–20% grain yield increases, hence enhance NUE under nitrogen-limiting conditions. Analysis of a panel of 2155 rice varieties reveals that 18% indica and 48% aus accessions carry small insertions in the ARE1 promoter, which result in a reduction in ARE1 expression and an increase in grain yield under nitrogen-limiting conditions. We propose that ARE1 is a key mediator of NUE and represents a promising target for breeding high-yield cultivars under nitrogen-limiting condition.
Abstract
Understanding the regulatory mechanisms of nitrogen assimilation is crucial for developing crop cultivars with improved nitrogen utilization efficiency (NUE). Here the authors identify a new negative regulator of NUE and mutation of this gene increases 10–20% rice grain yield under nitrogen-limiting field conditions.
Related collections
Most cited references27
- Record: found
- Abstract: found
- Article: not found
Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA.
- Record: found
- Abstract: found
- Article: not found
Plant nitrogen assimilation and use efficiency.
- Record: found
- Abstract: found
- Article: not found