- Record: found
- Abstract: found
- Article: found
The Drosophila homologue of CTIP1 (Bcl11a) and CTIP2 (Bcl11b) regulates neural stem cell temporal patterning
Read this article at
ABSTRACT
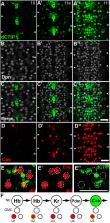
In the developing nervous system, neural stem cells (NSCs) use temporal patterning to generate a wide variety of different neuronal subtypes. In Drosophila, the temporal transcription factors, Hunchback, Kruppel, Pdm and Castor, are sequentially expressed by NSCs to regulate temporal identity during neurogenesis. Here, we identify a new temporal transcription factor that regulates the transition from the Pdm to Castor temporal windows. This factor, which we call Chronophage (or ‘time-eater’), is homologous to mammalian CTIP1 (Bcl11a) and CTIP2 (Bcl11b). We show that Chronophage binds upstream of the castor gene and regulates its expression. Consistent with Chronophage promoting a temporal switch, chronophage mutants generate an excess of Pdm-specified neurons and are delayed in generating neurons associated with the Castor temporal window. In addition to promoting the Pdm to Castor transition, Chronophage also represses the production of neurons generated during the earlier Hunchback and Kruppel temporal windows. Genetic interactions with Hunchback and Kruppel indicate that Chronophage regulates NSC competence to generate Hunchback- and Kruppel-specified neurons. Taken together, our results suggest that Chronophage has a conserved role in temporal patterning and neuronal subtype specification.
Abstract
Summary: The Drosophila CTIP1/2 (Bcl11a/b) homologue chronophage regulates the temporal cascade and neural stem cell competence in the embryonic central nervous system.
Related collections
Most cited references57
- Record: found
- Abstract: not found
- Article: not found
Enzymatic assembly of DNA molecules up to several hundred kilobases.
- Record: found
- Abstract: found
- Article: not found
An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases.
- Record: found
- Abstract: found
- Article: not found