- Record: found
- Abstract: found
- Article: found
A novel nonsense variant in TPM4 caused dominant macrothrombocytopenia, mild bleeding tendency and disrupted cytoskeleton remodeling
Read this article at
Abstract
Background
Rare inherited thrombocytopenias are caused by alterations in genes involved in megakaryopoiesis, thrombopoiesis and/or platelet release. Diagnosis is challenging due to poor specificity of platelet laboratory assays, large numbers of culprit genes, and difficult assessment of the pathogenicity of novel variants.
Objectives
To characterize the clinical and laboratory phenotype, and identifying the underlying molecular alteration, in a pedigree with thrombocytopenia of uncertain etiology.
Patients/Methods
Index case was enrolled in our Spanish multicentric project of inherited platelet disorders due to lifelong thrombocytopenia and bleeding. Bleeding score was recorded by ISTH‐BAT. Laboratory phenotyping consisted of blood cells count, blood film, platelet aggregation and flow cytometric analysis. Genotyping was made by whole‐exome sequencing (WES). Cytoskeleton proteins were analyzed in resting/spreading platelets by immunofluorescence and immunoblotting.
Results
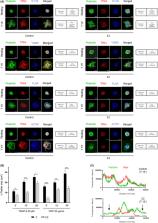
Five family members displayed lifelong mild thrombocytopenia with a high number of enlarged platelets in blood film, and mild bleeding tendency. Patient's platelets showed normal aggregation and granule secretion response to several agonists. WES revealed a novel nonsense variant (c.322C>T; p.Gln108*) in TPM4 (NM_003290.3), the gene encoding for tropomyosin‐4 (TPM4). This variant led to impairment of platelet spreading capacity after stimulation with TRAP‐6 and CRP, delocalization of TPM4 in activated platelets, and significantly reduced TPM4 levels in platelet lysates. Moreover, the index case displayed up‐regulation of TPM2 and TPM3 mRNA levels.
Related collections
Most cited references25

- Record: found
- Abstract: not found
- Article: not found