- Record: found
- Abstract: found
- Article: found
Clinical features and molecular genetic analysis in a Turkish family with oral white sponge nevus
Read this article at
Abstract
Background
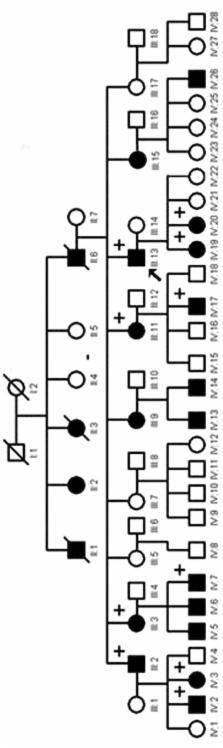
Oral white sponge nevus (WSN) is a rare autosomal dominant benign condition, characterized by asymptomatic spongy white plaques. Mutations in Keratin 4 (KRT4) and 13 (KRT13) have been shown to cause WSN. Familial cases are uncommon due to irregular penetrance. Thus, the aim of the study was: a) to demonstrate the clinical and histopathological features of a three-generation Turkish family with oral WSN b) to determine whether KRT4 or KRT13 gene mutation was the molecular basis of WSN.
Material and Methods
Out of twenty members of the family ten were available for assessment. Venous blood samples from six affected and five unaffected members and 48 healthy controls were obtained for genetic mutational analysis. Polymerase chain reaction was used to amplify all exons within KRT4 and KRT13 genes. These products were sequenced and the data was examined for mutations and polymorphisms.
Results
Varying presentation and severity of clinical features were observed. Analysis of the KRT13 gene revealed the sequence variant Y118D as the disease-causing mutation. One patient revealed several previously unreported polymorphisms including a novel mutation in exon 1 of the KRT13 gene and a heterozygous deletion in exon 1 of KRT4. This deletion in the KRT4 gene was found to be a common polymorphism reflecting a high allele frequency of 31.25% in the Turkish population.
Conclusions
Oral WSN may manifest variable clinical features. The novel mutation found in the KRT13 gene is believed to add evidence for a mutational hotspot in the mucosal keratins. Molecular genetic analysis is required to establish correct diagnosis and appropriate genetic consultation.
Key words:White sponge nevus, leukokeratosis, oral mucosa, keratins, mutation.
Related collections
Most cited references18
- Record: found
- Abstract: not found
- Article: not found
Intermediate filament proteins and their associated diseases.
- Record: found
- Abstract: found
- Article: not found
Human keratin diseases: the increasing spectrum of disease and subtlety of the phenotype-genotype correlation.
- Record: found
- Abstract: found
- Article: not found