- Record: found
- Abstract: found
- Article: found
High resolution cryo-EM structure of the helical RNA-bound Hantaan virus nucleocapsid reveals its assembly mechanisms
Read this article at
Abstract
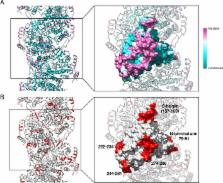
Negative-strand RNA viruses condense their genome into helical nucleocapsids that constitute essential templates for viral replication and transcription. The intrinsic flexibility of nucleocapsids usually prevents their full-length structural characterisation at high resolution. Here, we describe purification of full-length recombinant metastable helical nucleocapsid of Hantaan virus ( Hantaviridae family, Bunyavirales order) and determine its structure at 3.3 Å resolution by cryo-electron microscopy. The structure reveals the mechanisms of helical multimerisation via sub-domain exchanges between protomers and highlights nucleotide positions in a continuous positively charged groove compatible with viral genome binding. It uncovers key sites for future structure-based design of antivirals that are currently lacking to counteract life-threatening hantavirus infections. The structure also suggests a model of nucleoprotein-polymerase interaction that would enable replication and transcription solely upon local disruption of the nucleocapsid.
eLife digest
Rats and mice sometimes transmit hantaviruses, a family of microbes that can cause deadly human diseases. For example, the Hantaan virus leads to haemorrhagic fevers that are potentially fatal. There are no vaccine or even drugs against these infections.
To multiply, viruses must insert their genetic material inside a cell. While the body often detects and destroys foreign genetic information, hantaviruses can still evade our defences. Molecules called nucleoproteins bind to the viral genome, hiding it away in long helices called nucleocapsids. When the virus needs to replicate, an enzyme opens up the nucleocapsid, reads and copies the genetic code, and then closes the helix. Yet, researchers know little about the details of this process, or even the structure of the nucleocapsid.
Here, Arragain et al. use a method called cryo-electron microscopy to examine and piece together the exact 3D structure of the Hantaan virus nucleocapsid. This was possible because the new technique allows scientists to observe biological molecules at an unprecedented, near atomic resolution. The resulting model reveals that the viral genome nests into a groove inside the nucleocapsid. It also shows that specific interactions between nucleoproteins stabilise the helix. Finally, the model helps to provide hypotheses on how the enzyme could read the genome without breaking the capsid.
Mapping out the structure and the interactions of the nucleocapsid is the first step towards finding molecules that could destabilise the helix and neutralise the virus: this could help fight both the Hantaan virus and other members of its deadly family.
Related collections
Most cited references24
- Record: found
- Abstract: found
- Article: not found
Organization of the influenza virus replication machinery.
- Record: found
- Abstract: found
- Article: found
Structural Insights into Bunyavirus Replication and Its Regulation by the vRNA Promoter
- Record: found
- Abstract: found
- Article: not found