- Record: found
- Abstract: found
- Article: not found
First detection and molecular identification of Borrelia species in Bactrian camel ( Camelus bactrianus) from Northwest China
Abstract
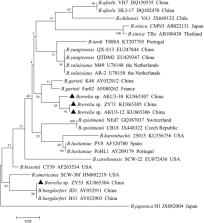
Comprehensive epidemiological surveys for Lyme disease have not been conducted for the Bactrian camel in China. In this study, a total of 138 blood specimens collected from Bactrian camels from Zhangye City in Gansu Province and Yili and Aksu in Xinjiang Province, China, were examined for the presence of Borrelia spp. Species-specificity nested PCR based on the 5S-23S rRNA, OspA, flaB and 16S rRNA genes revealed that the total positive rate of Borrelia spp. was 3.6% (5/138, 95% CI = 0.2–17.9). These results were confirmed by sequence analysis of the positive PCR products or positive colonies. This is the first report of Borrelia pathogens in camels in China. Two Borrelia species that cause Lyme disease and one that causes relapsing fever were identified in the camel blood samples by sequencing. The findings of this study indicate that the Bactrian camel may serve as a potential natural host of Lyme disease and/or relapsing fever in China.
Highlights
-
•
This is the first report of Borrelia pathogens using molecular tools in Bactrian camel.
-
•
Bactrian camel may play as potential natural host of Lyme disease in China.
-
•
The results revealed that the prevalence of Borrelia spp. were 3.6% based on sequences of four genes.
-
•
A novel Borrelia genespecies belong to Relapsing fever Borrelia spp. was found.
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: not found
Hosts as ecological traps for the vector of Lyme disease.
- Record: found
- Abstract: found
- Article: not found
Diversity of Borrelia burgdorferi sensu lato evidenced by restriction fragment length polymorphism of rrf (5S)-rrl (23S) intergenic spacer amplicons.
- Record: found
- Abstract: found
- Article: not found
