- Record: found
- Abstract: found
- Article: found
CSB-independent, XPC-dependent transcription-coupled repair in Drosophila
Read this article at
Significance
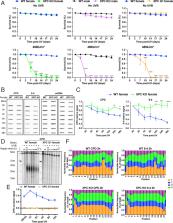
We have discovered that Drosophila, which does not have the canonical TCR homologs, does nevertheless carry out TCR as efficiently as organisms that do. Furthermore, using the XR-seq and in vivo excision assay we have also shown that both global repair and TCR in Drosophila are dependent on the XPC protein and in that regard, Drosophila excision repair is more similar to the monocellular eukaryotic yeast repair system than it is to multicellular eukaryotes. Finally, we have generated genome-wide single nucleotide repair maps of Drosophila for CPDs, (6-4) photoproducts, and cisplatin-d(GpG) adducts that should be a useful source for investigators working on DNA damage, repair, and mutagenesis in Drosophila.
Abstract
Drosophila melanogaster has been extensively used as a model system to study ionizing radiation and chemical-induced mutagenesis, double-strand break repair, and recombination. However, there are only limited studies on nucleotide excision repair in this important model organism. An early study reported that Drosophila lacks the transcription-coupled repair (TCR) form of nucleotide excision repair. This conclusion was seemingly supported by the Drosophila genome sequencing project, which revealed that Drosophila lacks a homolog to CSB, which is known to be required for TCR in mammals and yeasts. However, by using excision repair sequencing (XR-seq) genome-wide repair mapping technology, we recently found that the Drosophila S2 cell line performs TCR comparable to human cells. Here, we have extended this work to Drosophila at all its developmental stages. We find TCR takes place throughout the life cycle of the organism. Moreover, we find that in contrast to humans and other multicellular organisms previously studied, the XPC repair factor is required for both global and transcription-coupled repair in Drosophila.
Related collections
Most cited references51
- Record: found
- Abstract: found
- Article: not found
Transcription-coupled DNA repair: two decades of progress and surprises.
- Record: found
- Abstract: found
- Article: not found
RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo.
- Record: found
- Abstract: found
- Article: not found