- Record: found
- Abstract: found
- Article: found
Amblyomma imitator Ticks as Vectors of Rickettsia rickettsii, Mexico
brief-report
Karla A. Oliveira
1 ,
Adriano Pinter
2 ,
Aaron Medina-Sanchez
3 ,
Venkata D. Boppana ,
Stephen K. Wikel ,
Tais B. Saito ,
Thomas Shelite ,
Lucas Blanton ,
Vsevolod Popov ,
Pete D. Teel
4 ,
David H. Walker ,
Marcio A.M. Galvao
5 ,
Claudio Mafra
1 ,
Donald H. Bouyer
August 2010
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
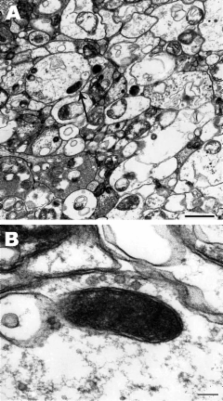
Real-time PCR of Amblyomma imitator tick egg masses obtained in Nuevo Leon State, Mexico, identified a Rickettsia species. Sequence analyses of 17-kD common antigen and outer membrane protein A and B gene fragments showed to it to be R. rickettsii, which suggested a potential new vector for this bacterium.
Related collections
Most cited references11
- Record: found
- Abstract: found
- Article: not found
Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes.
R Regnery, C L Spruill, B D Plikaytis (1991)
- Record: found
- Abstract: found
- Article: not found
Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rOmpB (ompB).
Ana V. Diez Roux, D Raoult (2000)
- Record: found
- Abstract: found
- Article: not found
Differentiation of spotted fever group rickettsiae by sequencing and analysis of restriction fragment length polymorphism of PCR-amplified DNA of the gene encoding the protein rOmpA.
Ana V. Diez Roux, P.-E. Fournier, D Raoult (1996)