- Record: found
- Abstract: found
- Article: not found
Dynein Modifiers in C. elegans: Light Chains Suppress Conditional Heavy Chain Mutants
Read this article at
Abstract
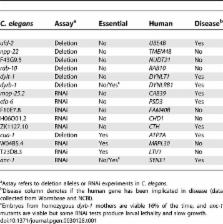
Cytoplasmic dynein is a microtubule-dependent motor protein that functions in mitotic cells during centrosome separation, metaphase chromosome congression, anaphase spindle elongation, and chromosome segregation. Dynein is also utilized during interphase for vesicle transport and organelle positioning. While numerous cellular processes require cytoplasmic dynein, the mechanisms that target and regulate this microtubule motor remain largely unknown. By screening a conditional Caenorhabditis elegans cytoplasmic dynein heavy chain mutant at a semipermissive temperature with a genome-wide RNA interference library to reduce gene functions, we have isolated and characterized twenty dynein-specific suppressor genes. When reduced in function, these genes suppress dynein mutants but not other conditionally mutant loci, and twelve of the 20 specific suppressors do not exhibit sterile or lethal phenotypes when their function is reduced in wild-type worms. Many of the suppressor proteins, including two dynein light chains, localize to subcellular sites that overlap with those reported by others for the dynein heavy chain. Furthermore, knocking down any one of four putative dynein accessory chains suppresses the conditional heavy chain mutants, suggesting that some accessory chains negatively regulate heavy chain function. We also identified 29 additional genes that, when reduced in function, suppress conditional mutations not only in dynein but also in loci required for unrelated essential processes. In conclusion, we have identified twenty genes that in many cases are not essential themselves but are conserved and when reduced in function can suppress conditionally lethal C. elegans cytoplasmic dynein heavy chain mutants. We conclude that conserved but nonessential genes contribute to dynein function during the essential process of mitosis.
Author Summary
Microtubules and microtubule-dependent motor proteins segregate chromosomes during mitosis and also promote cellular organization in nondividing cells. An essential motor protein complex called cytoplasmic dynein powers many aspects of microtubule-dependent transport, but it is currently unclear how dynein is regulated such that it can execute different processes. We have performed a genome-wide screen to isolate genes that are involved in dynein-dependent processes. We determined that 20 of the 49 genes we identified specifically influenced the viability of dynein mutant strains but not the viability of other C. elegans mutants. Many of the proteins that specifically influence dynein localized to subcellular sites where the dynein heavy chain has been reported by others to be found. Additionally, we identified four dynein components that appear to negatively regulate the force-generating dynein heavy chain. The identification and initial characterization of this group of genes represents a route to identify genes that are not themselves essential but do participate in essential processes.
Related collections
Most cited references67
- Record: found
- Abstract: found
- Article: not found
Functional genomic analysis of C. elegans chromosome I by systematic RNA interference.
- Record: found
- Abstract: found
- Article: not found
Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans.
- Record: found
- Abstract: found
- Article: not found
