- Record: found
- Abstract: found
- Article: found
The complete chloroplast genome of Lagerstroemia balansae, an endangered species of genus Lagerstroemia native to China
research-article
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
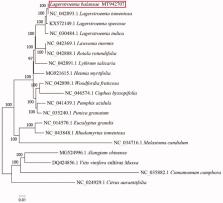
We announce here the first complete chloroplast genome sequence of Lagerstroemia balansae, a plant species with extremely small populations rated the level of EN (Endangered) in China. This complete chloroplast genome is 152316 bp in size. In total, 130 genes were identified, including 85 protein-coding genes, 37 tRNA genes, 8 rRNA genes. The result of phylogenetic analysis strongly supported that L. balansae was closely related to L. tomentosa.
Related collections
Most cited references10

- Record: found
- Abstract: found
- Article: found
MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability
Kazutaka Katoh, Daron Standley (2013)

- Record: found
- Abstract: found
- Article: found
IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies
Lam-Tung Nguyen, Heiko Schmidt, Arndt von Haeseler … (2014)
- Record: found
- Abstract: found
- Article: not found
ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates
Subha Kalyaanamoorthy, Bui Minh, Thomas Wong … (2017)