- Record: found
- Abstract: found
- Article: found
Protein Composition of the Bovine Herpesvirus 1.1 Virion
Read this article at
Abstract
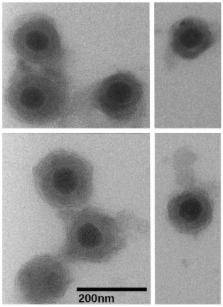
Bovine herpesvirus (BoHV) type 1 is an important agricultural pathogen that infects cattle and other ruminants worldwide. Acute infection of the oro-respiratory tract leads to immune suppression and allows commensal bacteria to infect an otherwise healthy lower respiratory tract. This condition is known as the Bovine Respiratory Disease (BRD). BoHV-1 latently infects the host for life and periodical stress events re-initiate BRD, translating into high morbidity and large economic losses. To gain a better understanding of the biology of BoHV-1 and the disease it causes, we elucidated the protein composition of extracellular virions using liquid chromatography-mass spectrometry analysis. We detected 33 viral proteins, including the expected proteins of the nucleocapsid and envelope as well as other regulatory proteins present in the viral tegument. In addition to viral proteins, we have also identified packaged proteins of host origin. This constitutes the first proteomic characterization of the BoHV virion.
Related collections
Most cited references77
- Record: found
- Abstract: found
- Article: not found
Bacterial pathogens of the bovine respiratory disease complex.
- Record: found
- Abstract: found
- Article: not found
Microtubule-mediated Transport of Incoming Herpes Simplex Virus 1 Capsids to the Nucleus
- Record: found
- Abstract: found
- Article: not found