- Record: found
- Abstract: found
- Article: found
Functional analysis of the BRI1 receptor kinase by Thr-for-Ser substitution in a regulatory autophosphorylation site
Read this article at
Abstract
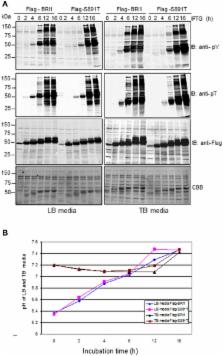
BRI1 becomes highly phosphorylated in vivo upon perception of the ligand, brassinolide, as a result of autophosphorylation and transphosphorylation by its co-receptor kinase, BAK1. Important autophosphorylation sites include those involved in activation of kinase activity and those that are inhibitory, such as Ser-891. The inhibitory sites are autophosphorylated after kinase activation has been achieved and are postulated to contribute to deactivation of the kinase. The function of phosphosites is usually tested by substituting a non-phosphorylatable residue or an acidic residue that can act as a phosphomimetic. What has typically not been examined is substitution of a Thr for a Ser phosphosite (or vice versa) but given that Thr and Ser are not equivalent amino acids this type of substitution may represent a new approach to engineer regulatory phosphorylation. In the present study with BRI1, we substituted Thr at the Ser-891 phosphosite to generate the S891T directed mutant. The recombinant Flag-BRI1 (S891T) cytoplasmic domain protein (the S891T protein) was catalytically active and phosphorylation occurred at the engineered Thr-891 site. However, the S891T recombinant protein autophosphorylated more slowly than the wild-type protein during expression in E. coli. As a result, activation of peptide kinase activity (measured in vitro) was delayed as was transphosphorylation of bacterial proteins in situ. Stable transgenic expression of BRI1 (S891T)-Flag in Arabidopsis bri1-5 plants did not fully rescue the brassinosteroid (BR) phenotype indicating that BR signaling was constrained. Our working model is that restricted signaling in the S891T plants occurs as a result of the reduced rate of activation of the mutant BRI1 kinase by autophosphorylation. These results provide the platform for future studies to critically test this new model in vivo and establish Ser-Thr substitutions at phosphosites as an interesting approach to consider with other protein kinases.
Related collections
Most cited references30
- Record: found
- Abstract: found
- Article: not found
Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases.
- Record: found
- Abstract: not found
- Article: not found
Active and Inactive Protein Kinases: Structural Basis for Regulation
- Record: found
- Abstract: found
- Article: not found