- Record: found
- Abstract: found
- Article: found
Mind the Outgroup and Bare Branches in Total-Evidence Dating: a Case Study of Pimpliform Darwin Wasps (Hymenoptera, Ichneumonidae)
Read this article at
Abstract
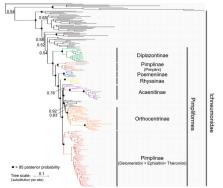
Taxon sampling is a central aspect of phylogenetic study design, but it has received limited attention in the context of total-evidence dating, a widely used dating approach that directly integrates molecular and morphological information from extant and fossil taxa. We here assess the impact of commonly employed outgroup sampling schemes and missing morphological data in extant taxa on age estimates in a total-evidence dating analysis under the uniform tree prior. Our study group is Pimpliformes, a highly diverse, rapidly radiating group of parasitoid wasps of the family Ichneumonidae. We analyze a data set comprising 201 extant and 79 fossil taxa, including the oldest fossils of the family from the Early Cretaceous and the first unequivocal representatives of extant subfamilies from the mid-Paleogene. Based on newly compiled molecular data from ten nuclear genes and a morphological matrix that includes 222 characters, we show that age estimates become both older and less precise with the inclusion of more distant and more poorly sampled outgroups. These outgroups not only lack morphological and temporal information but also sit on long terminal branches and considerably increase the evolutionary rate heterogeneity. In addition, we discover an artifact that might be detrimental for total-evidence dating: “bare-branch attraction,” namely high attachment probabilities of certain fossils to terminal branches for which morphological data are missing. Using computer simulations, we confirm the generality of this phenomenon and show that a large phylogenetic distance to any of the extant taxa, rather than just older age, increases the risk of a fossil being misplaced due to bare-branch attraction. After restricting outgroup sampling and adding morphological data for the previously attracting, bare branches, we recover a Jurassic origin for Pimpliformes and Ichneumonidae. This first age estimate for the group not only suggests an older origin than previously thought but also that diversification of the crown group happened well before the Cretaceous-Paleogene boundary. Our case study demonstrates that in order to obtain robust age estimates, total-evidence dating studies need to be based on a thorough and balanced sampling of both extant and fossil taxa, with the aim of minimizing evolutionary rate heterogeneity and missing morphological information. [Bare-branch attraction; ichneumonids; fossils; morphological matrix; phylogeny; RoguePlots.]
Related collections
Most cited references114

- Record: found
- Abstract: found
- Article: found
MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability
- Record: found
- Abstract: found
- Article: found
MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space

- Record: found
- Abstract: found
- Article: found