- Record: found
- Abstract: found
- Article: found
JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis
Read this article at
Significance
The plant hormone jasmonate promotes resistance to plant-eating organisms, ranging from pathogenic microbes to mammals. Jasmonate reprograms metabolism to fuel the production of diverse defense compounds and simultaneously inhibits plant growth. Understanding how growth is influenced across a range of defense levels remains unclear, but has important implications for optimizing crop productivity. Using a genetic approach to “tune” the jasmonate response, we assessed the physiological consequences of discrete levels of defense throughout the plant life cycle. Overactivation of jasmonate response led to carbon starvation, near loss of seed production and, under extreme conditions, lethality. Our findings explain the emergence of diverse strategies to keep jasmonate responses at bay and provide new insights into metabolic processes that underlie growth–defense trade-offs.
Abstract
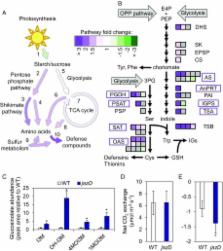
Plant immune responses mediated by the hormone jasmonoyl- l-isoleucine (JA-Ile) are metabolically costly and often linked to reduced growth. Although it is known that JA-Ile activates defense responses by triggering the degradation of JASMONATE ZIM DOMAIN (JAZ) transcriptional repressor proteins, expansion of the JAZ gene family in vascular plants has hampered efforts to understand how this hormone impacts growth and other physiological tasks over the course of ontogeny. Here, we combined mutations within the 13-member Arabidopsis JAZ gene family to investigate the effects of chronic JAZ deficiency on growth, defense, and reproductive output. A higher-order mutant ( jaz decuple, jazD) defective in 10 JAZ genes ( JAZ1– 7, -9, -10, and -13) exhibited robust resistance to insect herbivores and fungal pathogens, which was accompanied by slow vegetative growth and poor reproductive performance. Metabolic phenotypes of jazD discerned from global transcript and protein profiling were indicative of elevated carbon partitioning to amino acid-, protein-, and endoplasmic reticulum body-based defenses controlled by the JA-Ile and ethylene branches of immunity. Resource allocation to a strong defense sink in jazD leaves was associated with increased respiration and hallmarks of carbon starvation but no overt changes in photosynthetic rate. Depletion of the remaining JAZ repressors in jazD further exaggerated growth stunting, nearly abolished seed production and, under extreme conditions, caused spreading necrotic lesions and tissue death. Our results demonstrate that JAZ proteins promote growth and reproductive success at least in part by preventing catastrophic metabolic effects of an unrestrained immune response.
Related collections
Most cited references68
- Record: found
- Abstract: found
- Article: not found
JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling.
- Record: found
- Abstract: found
- Article: not found
JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis.
- Record: found
- Abstract: found
- Article: not found