- Record: found
- Abstract: found
- Article: found
Epigenetic regulation of rice flowering and reproduction
Read this article at
Abstract
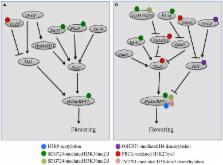
Current understanding of the epigenetic regulator roles in plant growth and development has largely derived from studies in the dicotyledonous model plant Arabidopsis thaliana. Rice ( Oryza sativa) is one of the most important food crops in the world and has more recently becoming a monocotyledonous model plant in functional genomics research. During the past few years, an increasing number of studies have reported the impact of DNA methylation, non-coding RNAs and histone modifications on transcription regulation, flowering time control, and reproduction in rice. Here, we review these studies to provide an updated complete view about chromatin modifiers characterized in rice and in particular on their roles in epigenetic regulation of flowering time, reproduction, and seed development.
Related collections
Most cited references104
- Record: found
- Abstract: found
- Article: not found
Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation.
- Record: found
- Abstract: found
- Article: not found