- Record: found
- Abstract: found
- Article: found
Spatial specificity of auxin responses coordinates wood formation
Read this article at
Abstract
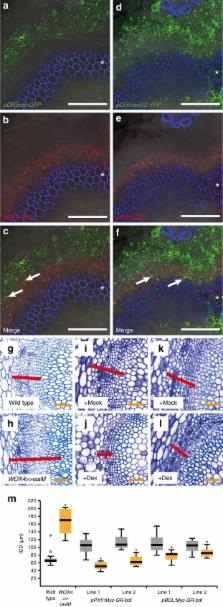
Spatial organization of signalling events of the phytohormone auxin is fundamental for maintaining a dynamic transition from plant stem cells to differentiated descendants. The cambium, the stem cell niche mediating wood formation, fundamentally depends on auxin signalling but its exact role and spatial organization is obscure. Here we show that, while auxin signalling levels increase in differentiating cambium descendants, a moderate level of signalling in cambial stem cells is essential for cambium activity. We identify the auxin-dependent transcription factor ARF5/MONOPTEROS to cell-autonomously restrict the number of stem cells by directly attenuating the activity of the stem cell-promoting WOX4 gene. In contrast, ARF3 and ARF4 function as cambium activators in a redundant fashion from outside of WOX4-expressing cells. Our results reveal an influence of auxin signalling on distinct cambium features by specific signalling components and allow the conceptual integration of plant stem cell systems with distinct anatomies.
Abstract
Auxin activity controls plant stem cell function. Here the authors show that in the cambium, moderate auxin activity restricts cambial stem cell number via ARF5-dependent repression of the stem‐cell‐promoting factor WOX4, while ARF3 and ARF4 promote cambial activity outside of the WOX4‐expression domain.
Related collections
Most cited references79
- Record: found
- Abstract: found
- Article: not found
Differential expression analysis for sequence count data

- Record: found
- Abstract: found
- Article: found
NCBI GEO: archive for functional genomics data sets—update
- Record: found
- Abstract: found
- Article: not found