- Record: found
- Abstract: found
- Article: not found
Hendra and Nipah viruses: different and dangerous
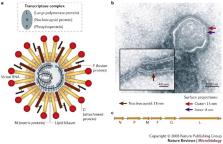
Key Points
-
The new genus Henipavirus was created within the Paramyxovirinae subfamily of the Paramyxoviridae for the Biosafety Level 4 (BSL4) pathogens Hendra virus and Nipah virus. Both are highly pathogenic paramyxoviruses that have recently emerged from flying foxes to cause serious disease outbreaks in humans and livestock in Australia, Malaysia, Singapore and Bangladesh.
-
Although they belong to the Paramyxovirinae subfamily, henipaviruses have distinct genetic and biological properties that distinguish them from other viruses in the subfamily, including respiroviruses such as Sendai virus, rubulaviruses such as mumps virus, and morbilliviruses such as measles virus.
-
Research on the henipaviruses has been restricted by their BSL4 categorization; however, recent results mainly obtained using henipavirus proteins expressed from cloned genes have increased our understanding of the unique properties of particular henipavirus proteins, particularly the attachment (G) protein, the fusion (F) protein and the phosphoprotein (P) gene products.
-
Among the features that distinguish the henipaviruses from other paramyxoviruses is their extraordinarily broad host range — they naturally infect flying foxes, horses, pigs, cats, dogs and humans — and the systemic infections that they cause, displaying a tropism for arterial rather than venous endothelial cells. The recent identification of the membrane receptor for the henipavirus G protein could explain these observations. The G protein of both HeV and NiV binds to ephrin B2, a conserved cell-surface glycoprotein that is widely distributed in vertebrates and is located preferentially in arterial endothelial cells and the surrounding tunica media, but is not found in venous endothelial cells. Ephrin B2 is also found in neurons, providing an explanation for virus growth in brain tissue and the occurrence of encephalitis in human patients.
-
The F protein is a type I membrane protein, and a biologically active form of the F protein is generated by the proteolytic cleavage of a protein precursor. It was recently found that the henipavirus F protein is cleaved by the endosomal protease cathepsin L, at a cleavage site that is unique among viral glycoproteins. The widespread distribution of the cathepsin L might also be crucial in the systemic spread of virus and the transmission of infectious virus within and between species.
-
The paramyxovirus P gene encodes three transcripts: the P, V and W proteins, each of which has a unique C-terminal domain and, compared with morbilliviruses and rubulaviruses, an N-terminal extension of 100–200 amino acids. P-gene products allow henipaviruses to evade host antiviral defences by inhibiting both dsRNA signalling and interferon (IFN) signalling. Both the V and W proteins inhibit dsRNA signalling, but their distinct C-terminal domains enable them to do so in different cellular compartments; the W protein contains a nuclear-localization signal in the C-terminal domain. The P, V and W proteins also inhibit IFN signalling by targeting the STAT proteins in a novel strategy for paramyxoviruses that involves STATs being sequestered in high-molecular-weight complexes and, again, the W protein acts in the nucleus.
Abstract
The highly virulent paramyxoviruses Hendra and Nipah virus are recent additions to the gamut of emerging human pathogens. Bryan Eaton and colleagues provide an overview of these pathogens and discuss recent progress in the understanding of the molecular basis for henipavirus pathogenicity.
Abstract
Hendra virus and Nipah virus are highly pathogenic paramyxoviruses that have recently emerged from flying foxes to cause serious disease outbreaks in humans and livestock in Australia, Malaysia, Singapore and Bangladesh. Their unique genetic constitution, high virulence and wide host range set them apart from other paramyxoviruses. These features led to their classification into the new genus Henipavirus within the family Paramyxoviridae and to their designation as Biosafety Level 4 pathogens. This review provides an overview of henipaviruses and the types of infection they cause, and describes how studies on the structure and function of henipavirus proteins expressed from cloned genes have provided insights into the unique biological properties of these emerging human pathogens.
Related collections
Most cited references135
- Record: found
- Abstract: found
- Article: not found
Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats.
- Record: found
- Abstract: found
- Article: not found
Triggering the interferon antiviral response through an IKK-related pathway.
Author and article information
Comments
Comment on this article
 Smart Citations
Smart CitationsSee how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.