- Record: found
- Abstract: found
- Article: found
DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation
Read this article at
Abstract
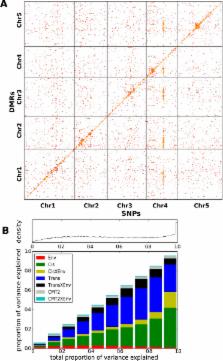
Epigenome modulation potentially provides a mechanism for organisms to adapt, within and between generations. However, neither the extent to which this occurs, nor the mechanisms involved are known. Here we investigate DNA methylation variation in Swedish Arabidopsis thaliana accessions grown at two different temperatures. Environmental effects were limited to transposons, where CHH methylation was found to increase with temperature. Genome-wide association studies (GWAS) revealed that the extensive CHH methylation variation was strongly associated with genetic variants in both cis and trans, including a major trans-association close to the DNA methyltransferase CMT2. Unlike CHH methylation, CpG gene body methylation (GBM) was not affected by growth temperature, but was instead correlated with the latitude of origin. Accessions from colder regions had higher levels of GBM for a significant fraction of the genome, and this was associated with increased transcription for the genes affected. GWAS revealed that this effect was largely due to trans-acting loci, many of which showed evidence of local adaptation.
eLife digest
Organisms need to adapt quickly to changes in their environment. Mutations in the DNA sequence of genes can lead to new adaptations, but this can take many generations. Instead, altering how genes are switched on by changing how the DNA is packaged in cells can allow organisms to adapt within and between generations. One way that genes are controlled in organisms is by a process known as DNA methylation, where ‘methyl’ tags are added to DNA and act as markers for other proteins involved in activating genes.
DNA is made of four different molecules called ‘nucleotides’ that are arranged in different orders to produce a vast variety of DNA sequences. One type of DNA methylation can happen at sites where a nucleotide called cytosine is followed by two other non-cytosine nucleotides. Another type of methylation can take place at sites where a cytosine is followed by a guanine nucleotide. However, it is not clear how big a role DNA methylation plays in allowing organisms to adapt to their changing environment.
Here, Dubin, Zhang, Meng, Remigereau et al. studied DNA methylation in a plant called Arabidopsis thaliana. Several different varieties of A. thaliana plants from Sweden were grown at two different temperatures. The experiments showed that the A. thaliana plants grown at higher temperatures were more likely to have methyl tags attached to sections of DNA called transposons, which are able to move around the genome. There was a lot of variety in the levels of this DNA methylation in the different plants, and some of it was shown to be associated with variation in a gene that is involved in DNA methylation.
However, not all of the DNA methylation in these plants was sensitive to the temperature the plants were grown in. Dubin, Zhang, Meng, Remigereau et al. show that the pattern of a type of DNA methylation that is found within genes depends on how far north in Sweden the plants' ancestors came from rather than the temperature the plants were grown in. Plants that originated from colder regions, farther north, had more DNA methylation within many genes and these genes were more active.
These findings suggest that genetic differences in these plants strongly influence the levels of DNA methylation, and they provide the first direct link between DNA methylation and adaption to the environment. Future studies should reveal how DNA methylation is regulated in these plants, and whether it plays a key role in adaptation, or merely reflects other changes in the genome.
Related collections
Most cited references25
- Record: found
- Abstract: found
- Article: not found
An efficient multi-locus mixed model approach for genome-wide association studies in structured populations

- Record: found
- Abstract: found
- Article: found
Estimating and interpreting F ST: The impact of rare variants
- Record: found
- Abstract: found
- Article: not found