- Record: found
- Abstract: found
- Article: found
Alternative Strategies for Microbial Remediation of Pollutants via Synthetic Biology
Read this article at
Abstract
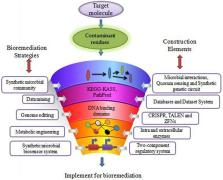
Continuous contamination of the environment with xenobiotics and related recalcitrant compounds has emerged as a serious pollution threat. Bioremediation is the key to eliminating persistent contaminants from the environment. Traditional bioremediation processes show limitations, therefore it is necessary to discover new bioremediation technologies for better results. In this review we provide an outlook of alternative strategies for bioremediation via synthetic biology, including exploring the prerequisites for analysis of research data for developing synthetic biological models of microbial bioremediation. Moreover, cell coordination in synthetic microbial community, cell signaling, and quorum sensing as engineered for enhanced bioremediation strategies are described, along with promising gene editing tools for obtaining the host with target gene sequences responsible for the degradation of recalcitrant compounds. The synthetic genetic circuit and two-component regulatory system (TCRS)-based microbial biosensors for detection and bioremediation are also briefly explained. These developments are expected to increase the efficiency of bioremediation strategies for best results.
Related collections
Most cited references193
- Record: found
- Abstract: found
- Article: not found
Modeling and simulation of genetic regulatory systems: a literature review.
- Record: found
- Abstract: found
- Article: not found
