- Record: found
- Abstract: found
- Article: found
Integration of Brain and Skull in Prenatal Mouse Models of Apert and Crouzon Syndromes
Read this article at
Abstract
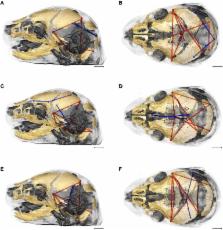
The brain and skull represent a complex arrangement of integrated anatomical structures composed of various cell and tissue types that maintain structural and functional association throughout development. Morphological integration, a concept developed in vertebrate morphology and evolutionary biology, describes the coordinated variation of functionally and developmentally related traits of organisms. Syndromic craniosynostosis is characterized by distinctive changes in skull morphology and perceptible, though less well studied, changes in brain structure and morphology. Using mouse models for craniosynostosis conditions, our group has precisely defined how unique craniosynostosis causing mutations in fibroblast growth factor receptors affect brain and skull morphology and dysgenesis involving coordinated tissue-specific effects of these mutations. Here we examine integration of brain and skull in two mouse models for craniosynostosis: one carrying the FGFR2c C342Y mutation associated with Pfeiffer and Crouzon syndromes and a mouse model carrying the FGFR2 S252W mutation, one of two mutations responsible for two-thirds of Apert syndrome cases. Using linear distances estimated from three-dimensional coordinates of landmarks acquired from dual modality imaging of skull (high resolution micro-computed tomography and magnetic resonance microscopy) of mice at embryonic day 17.5, we confirm variation in brain and skull morphology in Fgfr2c C342Y/+ mice, Fgfr2 +/ S252W mice, and their unaffected littermates. Mutation-specific variation in neural and cranial tissue notwithstanding, patterns of integration of brain and skull differed only subtly between mice carrying either the FGFR2c C342Y or the FGFR2 S252W mutation and their unaffected littermates. However, statistically significant and substantial differences in morphological integration of brain and skull were revealed between the two mutant mouse models, each maintained on a different strain. Relative to the effects of disease-associated mutations, our results reveal a stronger influence of the background genome on patterns of brain-skull integration and suggest robust genetic, developmental, and evolutionary relationships between neural and skeletal tissues of the head.
Related collections
Most cited references84
- Record: found
- Abstract: not found
- Article: not found
Morphological Integration and Developmental Modularity
- Record: found
- Abstract: not found
- Article: not found
FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease.
- Record: found
- Abstract: found
- Article: not found