- Record: found
- Abstract: found
- Article: found
Exploring the diversity of virulence genes in the Magnaporthe population infecting millets and rice in India
Read this article at
Abstract
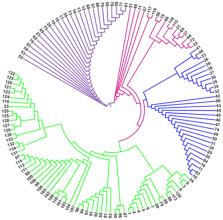
Blast pathogen, Magnaporthe spp., that infects ancient millet crops such pearl millet, finger millet, foxtail millet, barnyard millet, and rice was isolated from different locations of blast hotspots in India using single spore isolation technique and 136 pure isolates were established. Numerous growth characteristics were captured via morphogenesis analysis. Among the 10 investigated virulent genes, we could amplify MPS1 (TTK Protein Kinase) and Mlc (Myosin Regulatory Light Chain edc4) in majority of tested isolates, regardless of the crop and region where they were collected, indicating that these may be crucial for their virulence. Additionally, among the four avirulence ( Avr) genes studied, Avr-Pizt had the highest frequency of occurrence, followed by Avr-Pia. It is noteworthy to mention that Avr-Pik was present in the least number of isolates (9) and was completely absent from the blast isolates from finger millet, foxtail millet, and barnyard millet. A comparison at the molecular level between virulent and avirulent isolates indicated observably large variation both across (44%) and within (56%) them. The 136 Magnaporthe spp isolates were divided into four groups using molecular markers. Regardless of their geographic distribution, host plants, or tissues affected, the data indicate that the prevalence of numerous pathotypes and virulence factors at the field level, which may lead to a high degree of pathogenic variation. This research could be used for the strategic deployment of resistant genes to develop blast disease-resistant cultivars in rice, pearl millet, finger millet, foxtail millet, and barnyard millet.
Related collections
Most cited references57
- Record: found
- Abstract: found
- Article: not found
Detecting the number of clusters of individuals using the software structure: a simulation study
- Record: found
- Abstract: found
- Article: not found
Inference of Population Structure Using Multilocus Genotype Data
- Record: found
- Abstract: not found
- Article: not found