- Record: found
- Abstract: found
- Article: found
Rapid Differential Detection of Abrin Isoforms by an Acetonitrile- and Ultrasound-Assisted On-Bead Trypsin Digestion Coupled with LC-MS/MS Analysis
Read this article at
Abstract
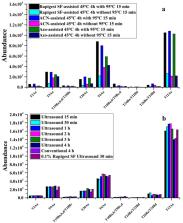
The high toxic abrin from the plant Abrus precatorius is a type II ribosome-inactivating protein toxin with a human lethal dose of 0.1–1.0 µg/kg body weight. Due to its high toxicity and the potential misuse as a biothreat agent, it is of great importance to developing fast and reliable methods for the identification and quantification of abrin in complex matrices. Here, we report rapid and efficient acetonitrile (ACN)- and ultrasound-assisted on-bead trypsin digestion method combined with HPLC-MS/MS for the quantification of abrin isoforms in complex matrices. Specific peptides of abrin isoforms were generated by direct ACN-assisted trypsin digestion and analyzed by HPLC-HRMS. Combined with in silico digestion and BLASTp database search, fifteen marker peptides were selected for differential detection of abrin isoforms. The abrin in milk and plasma was enriched by immunomagnetic beads prepared by biotinylated anti-abrin polyclonal antibodies conjugated to streptavidin magnetic beads. The ultrasound-assisted on-bead trypsin digestion method was carried out under the condition of 10% ACN as denaturant solvent, the entire digestion time was further shortened from 90 min to 30 min. The four peptides of T3A a,b,c,d, T12A a, T15A b, and T9A c,d were chosen as quantification for total abrin, abrin-a, abrin-b, and abrin-c/d, respectively. The absolute quantification of abrin and its isoforms was accomplished by isotope dilution with labeled AQUA peptides and analyzed by HPLC-MS/MS (MRM). The developed method was fully validated in milk and plasma matrices with quantification limits in the range of 1.0-9.4 ng/mL for the isoforms of abrin. Furthermore, the developed approach was applied for the characterization of abrin isoforms from various fractions from gel filtration separation of the seeds, and measurement of abrin in the samples of biotoxin exercises organized by the Organization for the Prohibition of Chemical Weapons (OPCW). This study provided a recommended method for the differential identification of abrin isoforms, which are easily applied in international laboratories to improve the capabilities for the analysis of biotoxin samples.
Related collections
Most cited references26
- Record: found
- Abstract: found
- Article: not found
A photo-cleavable surfactant for top-down proteomics
- Record: found
- Abstract: not found
- Article: not found