- Record: found
- Abstract: found
- Article: found
Analyzing the multi-target pharmacological mechanism of folium Artemisia argyi acting on breast cancer: a network pharmacology approach
Read this article at
Abstract
Background
Folium Artemisia argyi (FAA) is a traditional Chinese herbal medicine that is widely used in the clinic. However, the underlying mechanisms of its anticancer effects have not been fully elucidated.
Methods
In this study, we applied a network pharmacology approach to identify the potential mechanisms of FAA against breast cancer. To be specific, we screened the active ingredients and potential targets of the FAA through the Traditional Chinese Medicine Systems Pharmacology (TCMSP) database. Meanwhile, we employed the oral bioavailability (OB) and drug-likeness (DL) to search for potential bioactive compounds of FAA. Breast cancer-related target genes data were gathered from the GeneCards and Online Mendelian Inheritance in Man (OMIM) databases, and the protein-protein interaction (PPI) data were acquired from the Search Tool for the Retrieval of Interacting Genes (STRING) database. In addition, we constructed the network and performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis.
Results
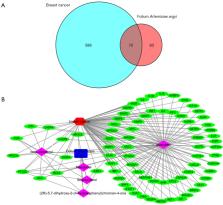
We obtained a total of nine active ingredients and 236 potential targets from FAA to construct a network, which showed that quercetin served as the major ingredient in FAA. AKT1 (RAC-alpha serine/threonine-protein kinase), MYC (Myc proto-oncogene protein), CASP3 (Caspase-3), EGFR (Epidermal growth factor receptor), JUN (Transcription factor AP-1), CCND1 (G1/S-specific cyclin-D1), VEGFA (Vascular endothelial growth factor A), ESR1 (Estrogen receptor), MAPK1 (Mitogen-activated protein kinase 1), and EGF (pro-epidermal growth factor) were identified as key targets of FAA in the treatment of breast cancer. The PPI cluster demonstrated that AKT1 was the seed in this cluster, indicating that AKT1 played a crucial role in connecting other nodes in the PPI network. This enrichment demonstrated that FAA was highly related to signal transduction, endocrine system, replication and repair, as well as cell growth and death. The enrichment results also verified that the underlying mechanisms of FAA against breast cancer might be attributed to the coordinated regulation of several cancer-related pathways, such as the MAPK and mammalian target of rapamycin (mTOR) signaling pathways, among others.
Related collections
Most cited references65
- Record: found
- Abstract: found
- Article: not found
Gene Ontology: tool for the unification of biology
- Record: found
- Abstract: found
- Article: not found
Cytoscape: a software environment for integrated models of biomolecular interaction networks.
- Record: found
- Abstract: found
- Article: not found