- Record: found
- Abstract: found
- Article: found
Improving the efficiency and accuracy of cardiovascular magnetic resonance with artificial intelligence—review of evidence and proposition of a roadmap to clinical translation

Read this article at
Abstract
Background
Cardiovascular magnetic resonance (CMR) is an important imaging modality for the assessment of heart disease; however, limitations of CMR include long exam times and high complexity compared to other cardiac imaging modalities. Recently advancements in artificial intelligence (AI) technology have shown great potential to address many CMR limitations. While the developments are remarkable, translation of AI-based methods into real-world CMR clinical practice remains at a nascent stage and much work lies ahead to realize the full potential of AI for CMR.
Methods
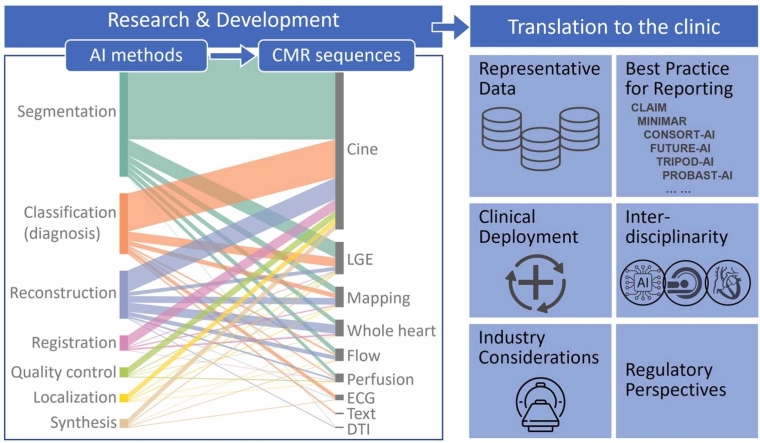
Herein we review recent cutting-edge and representative examples demonstrating how AI can advance CMR in areas such as exam planning, accelerated image reconstruction, post-processing, quality control, classification and diagnosis.
Results
These advances can be applied to speed up and simplify essentially every application including cine, strain, late gadolinium enhancement, parametric mapping, 3D whole heart, flow, perfusion and others. AI is a unique technology based on training models using data. Beyond reviewing the literature, this paper discusses important AI-specific issues in the context of CMR, including (1) properties and characteristics of datasets for training and validation, (2) previously published guidelines for reporting CMR AI research, (3) considerations around clinical deployment, (4) responsibilities of clinicians and the need for multi-disciplinary teams in the development and deployment of AI in CMR, (5) industry considerations, and (6) regulatory perspectives.
Graphical abstract
Related collections
Most cited references237
- Record: found
- Abstract: found
- Article: found
The FAIR Guiding Principles for scientific data management and stewardship
- Record: found
- Abstract: found
- Article: not found
nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation
- Record: found
- Abstract: found
- Article: not found