- Record: found
- Abstract: found
- Article: found
Genome‐wide association study for 13 agronomic traits reveals distribution of superior alleles in bread wheat from the Yellow and Huai Valley of China
Read this article at
Summary
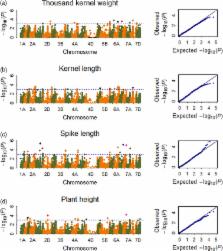
Bread wheat is a leading cereal crop worldwide. Limited amount of superior allele loci restricted the progress of molecular improvement in wheat breeding. Here, we revealed new allelic variation distribution for 13 yield‐related traits in series of genome‐wide association studies ( GWAS) using the wheat 90K genotyping assay, characterized in 163 bread wheat cultivars. Agronomic traits were investigated in 14 environments at three locations over 3 years. After filtering SNP data sets, GWAS using 20 689 high‐quality SNPs associated 1769 significant loci that explained, on average, ~20% of the phenotypic variation, both detected already reported loci and new promising genomic regions. Of these, repetitive and pleiotropic SNPs on chromosomes 6 AS, 6 AL, 6 BS, 5 BL and 7 AS were significantly linked to thousand kernel weight, for example BS00021705_51 on 6 BS and wsnp_Ex_c32624_41252144 on 6 AS, with phenotypic variation explained ( PVE) of ~24%, consistently identified in 12 and 13 of the 14 environments, respectively. Kernel length‐related SNPs were mainly identified on chromosomes 7 BS, 6 AS, 5 AL and 5 BL. Plant height‐related SNPs on chromosomes 4 DS, 6 DL, 2 DS and 1 BL were, respectively, identified in more than 11 environments, with averaged PVE of ~55%. Four SNPs were confirmed to be important genetic loci in two RIL populations. Based on repetivity and PVE, a total of 41 SNP loci possibly played the key role in modulating yield‐related traits of the cultivars surveyed. Distribution of superior alleles at the 41 SNP loci indicated that superior alleles were getting popular with time and modern cultivars had integrated many superior alleles, especially for peduncle length‐ and plant height‐related superior alleles. However, there were still 19 SNP loci showing less than percentages of 50% in modern cultivars, suggesting they should be paid more attention to improve yield‐related traits of cultivars in the Yellow and Huai wheat region. This study could provide useful information for dissection of yield‐related traits and valuable genetic loci for marker‐assisted selection in Chinese wheat breeding programme.
Related collections
Most cited references36
- Record: found
- Abstract: found
- Article: not found
A modified algorithm for the improvement of composite interval mapping.
- Record: found
- Abstract: found
- Article: not found
Population stratification and spurious allelic association.
- Record: found
- Abstract: found
- Article: not found