- Record: found
- Abstract: found
- Article: found
Reduced protein-coding transcript diversity in severe dengue emphasises the role of alternative splicing

Read this article at
Abstract
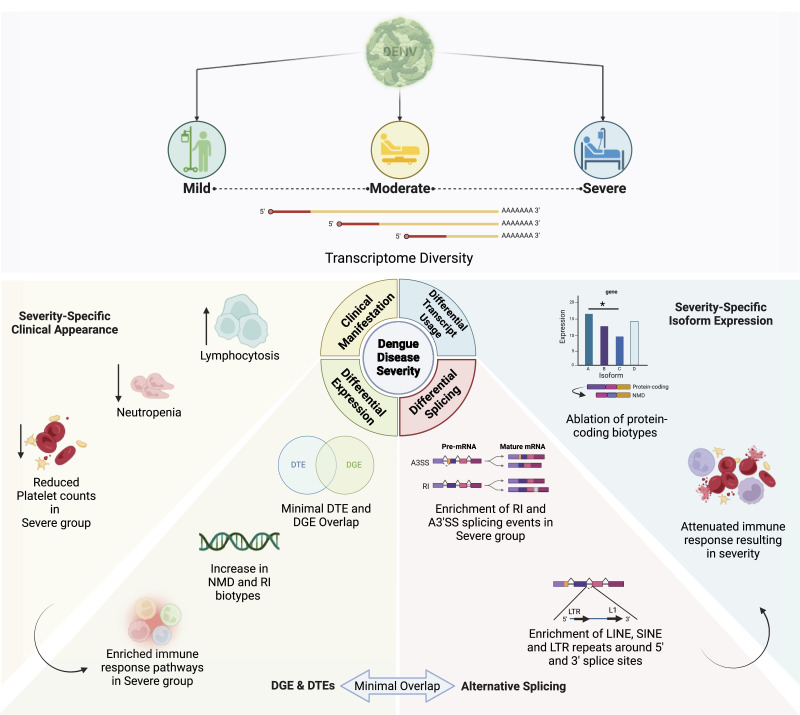
Transcriptomic analysis of dengue-infected patients reveals altered immune response pathways, transcript diversity, and splicing efficiency, underscoring potential therapeutic targets for treatment.
Abstract
Dengue fever, a neglected tropical arboviral disease, has emerged as a global health concern in the past decade. Necessitating a nuanced comprehension of the intricate dynamics of host–virus interactions influencing disease severity, we analysed transcriptomic patterns using bulk RNA-seq from 112 age- and gender-matched NS1 antigen–confirmed hospital-admitted dengue patients with varying severity. Severe cases exhibited reduced platelet count, increased lymphocytosis, and neutropenia, indicating a dysregulated immune response. Using bulk RNA-seq, our analysis revealed a minimal overlap between the differentially expressed gene and transcript isoform, with a distinct expression pattern across the disease severity. Severe patients showed enrichment in retained intron and nonsense-mediated decay transcript biotypes, suggesting altered splicing efficiency. Furthermore, an up-regulated programmed cell death, a haemolytic response, and an impaired interferon and antiviral response at the transcript level were observed. We also identified the potential involvement of the RBM39 gene among others in the innate immune response during dengue viral pathogenesis, warranting further investigation. These findings provide valuable insights into potential therapeutic targets, underscoring the importance of exploring transcriptomic landscapes between different disease sub-phenotypes in infectious diseases.
Graphical Abstract
Related collections
Most cited references86

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data

- Record: found
- Abstract: found
- Article: found