- Record: found
- Abstract: found
- Article: found
Panobinostat enhances olaparib efficacy by modifying expression of homologous recombination repair and immune transcripts in ovarian cancer
Read this article at
Highlights
-
•
Panobinostat in combination with olaparib reduces homologous recombination (HR) proficient tumor cell viability.
-
•
This combination favors anti-tumorigenic macrophages and enhances CD8+ T cell infiltration into tumors.
-
•
As a result, this combination targets HR-proficient ovarian tumors directly and through the tumor microenvironment.
Abstract
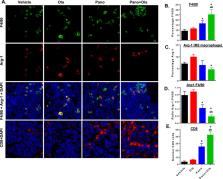
Histone deacetylase inhibitors (HDACi) sensitize homologous recombination (HR)-proficient human ovarian cancer cells to PARP inhibitors (PARPi). To investigate mechanisms of anti-tumor effects of combined HDACi/PARPi treatment we performed transcriptome analysis in HR- proficient human ovarian cancer cells and tested drug effects in established immunocompetent mouse ovarian cancer models. Human SKOV-3 cells were treated with vehicle (Con), olaparib (Ola), panobinostat (Pano) or Pano+Ola and RNA-seq analysis performed. DESeq2 identified differentially expressed HR repair and immune transcripts. Luciferised syngeneic mouse ovarian cancer cells (ID8-luc) were treated with the HDACi panobinostat alone or combined with olaparib and effects on cell viability, apoptosis, DNA damage and HR efficiency determined. C57BL/6 mice with intraperitoneally injected ID8-luc cells were treated with panobinostat and/or olaparib followed by assessment of tumor burden, markers of cell proliferation, apoptosis and DNA damage, tumor-infiltrating T cells and macrophages, and other immune cell populations in ascites fluid. There was a significant reduction in expression of 20/37 HR pathway genes by Pano+Ola, with immune and inflammatory-related pathways also significantly enriched by the combination. In ID8 cells, Pano+Ola decreased cell viability, HR repair, and enhanced DNA damage. Pano+Ola also co-operatively reduced tumor burden and proliferation, increased tumor apoptosis and DNA damage, enhanced infiltration of CD8+ T cells into tumors, and decreased expression of M2-like macrophage markers. In conclusion , panobinostat in combination with olaparib targets ovarian tumors through both direct cytotoxic and indirect immune-modulating effects.
Related collections
Most cited references48

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
Cancer Statistics, 2021
- Record: found
- Abstract: found
- Article: not found