- Record: found
- Abstract: found
- Article: found
Machine learning in time-lapse imaging to differentiate embryos from young vs old mice †

Read this article at
Abstract
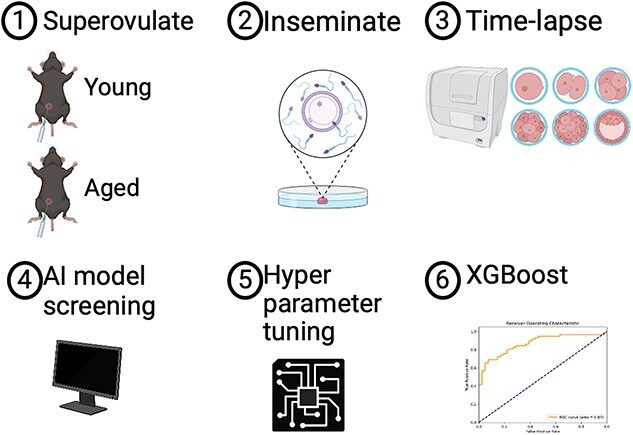
Time-lapse microscopy for embryos is a non-invasive technology used to characterize early embryo development. This study employs time-lapse microscopy and machine learning to elucidate changes in embryonic growth kinetics with maternal aging. We analyzed morphokinetic parameters of embryos from young and aged C57BL6/NJ mice via continuous imaging. Our findings show that aged embryos accelerated through cleavage stages (from 5-cells) to morula compared to younger counterparts, with no significant differences observed in later stages of blastulation. Unsupervised machine learning identified two distinct clusters comprising of embryos from aged or young donors. Moreover, in supervised learning, the extreme gradient boosting algorithm successfully predicted the age-related phenotype with 0.78 accuracy, 0.81 precision, and 0.83 recall following hyperparameter tuning. These results highlight two main scientific insights: maternal aging affects embryonic development pace, and artificial intelligence can differentiate between embryos from aged and young maternal mice by a non-invasive approach. Thus, machine learning can be used to identify morphokinetics phenotypes for further studies. This study has potential for future applications in selecting human embryos for embryo transfer, without or in complement with preimplantation genetic testing.
Abstract
Maternal aging in murine in vitro fertilization shows faster cleavage and compaction stages of embryonic development by time-lapse microscopy, and artificial intelligence predicts the aging phenotype.
Graphical Abstract
Related collections
Most cited references108
- Record: found
- Abstract: not found
- Article: not found
Scikit-Learn: Machine Learning in Python
- Record: found
- Abstract: found
- Article: not found
A guide to machine learning for biologists
- Record: found
- Abstract: found
- Article: not found