- Record: found
- Abstract: found
- Article: found
Design of protein-binding peptides with controlled binding affinity: the case of SARS-CoV-2 receptor binding domain and angiotensin-converting enzyme 2 derived peptides
Read this article at
Abstract
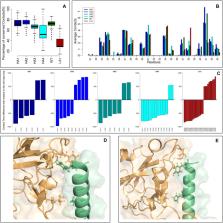
The development of methods able to modulate the binding affinity between proteins and peptides is of paramount biotechnological interest in view of a vast range of applications that imply designed polypeptides capable to impair or favour Protein-Protein Interactions. Here, we applied a peptide design algorithm based on shape complementarity optimization and electrostatic compatibility and provided the first experimental in vitro proof of the efficacy of the design algorithm. Focusing on the interaction between the SARS-CoV-2 Spike Receptor-Binding Domain (RBD) and the human angiotensin-converting enzyme 2 (ACE2) receptor, we extracted a 23-residues long peptide that structurally mimics the major interacting portion of the ACE2 receptor and designed in silico five mutants of such a peptide with a modulated affinity. Remarkably, experimental K D measurements, conducted using biolayer interferometry, matched the in silico predictions. Moreover, we investigated the molecular determinants that govern the variation in binding affinity through molecular dynamics simulation, by identifying the mechanisms driving the different values of binding affinity at a single residue level. Finally, the peptide sequence with the highest affinity, in comparison with the wild type peptide, was expressed as a fusion protein with human H ferritin (HFt) 24-mer. Solution measurements performed on the latter constructs confirmed that peptides still exhibited the expected trend, thereby enhancing their efficacy in RBD binding. Altogether, these results indicate the high potentiality of this general method in developing potent high-affinity vectors for hindering/enhancing protein-protein associations.
Related collections
Most cited references71
- Record: found
- Abstract: found
- Article: not found
Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein
- Record: found
- Abstract: not found
- Article: not found