- Record: found
- Abstract: found
- Article: not found
Structure-based drug repositioning over the human TMPRSS2 protease domain: search for chemical probes able to repress SARS-CoV-2 Spike protein cleavages
Read this article at
Abstract
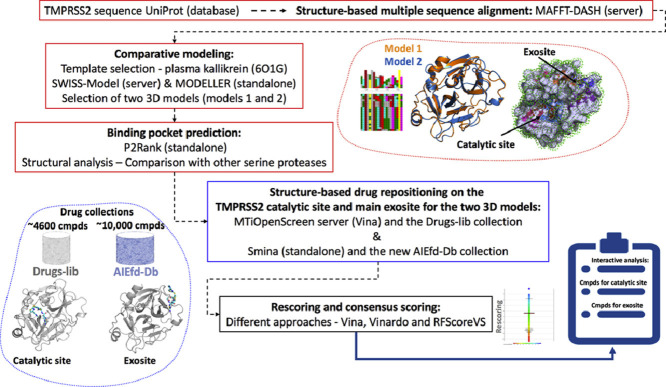
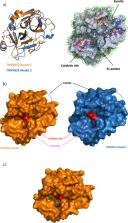
In December 2019, a new coronavirus was identified in the Hubei province of central china and named SARS-CoV-2. This new virus induces COVID-19, a severe respiratory disease with high death rate. A putative target to interfere with the virus is the host transmembrane serine protease family member II (TMPRSS2). This enzyme is critical for the entry of coronaviruses into human cells by cleaving and activating the spike protein (S) of SARS-CoV-2. Repositioning approved, investigational and experimental drugs on the serine protease domain of TMPRSS2 could thus be valuable. There is no experimental structure for TMPRSS2 but it is possible to develop quality structural models for the serine protease domain using comparative modeling strategies as such domains are highly structurally conserved. Beside the TMPRSS2 catalytic site, we predicted on our structural models a main exosite that could be important for the binding of protein partners and/or substrates. To block the catalytic site or the exosite of TMPRSS2 we used structure-based virtual screening computations and two different collections of approved, investigational and experimental drugs. We propose a list of 156 molecules that could bind to the catalytic site and 100 compounds that may interact with the exosite. These small molecules should now be tested in vitro to gain novel insights over the roles of TMPRSS2 or as starting point for the development of second generation analogs.
Graphical abstract
Related collections
Most cited references55

- Record: found
- Abstract: found
- Article: found
A pneumonia outbreak associated with a new coronavirus of probable bat origin
- Record: found
- Abstract: found
- Article: not found