- Record: found
- Abstract: found
- Article: found
Ethylene‐responsive factor ERF114 mediates fungal pathogen effector PevD1‐induced disease resistance in Arabidopsis thaliana
Read this article at
Abstract
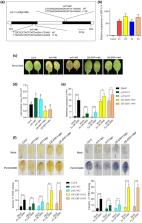
APETALA2/ethylene‐responsive factor (AP2/ERF) family transcription factors are well‐documented in plant responses to a wide range of biotic and abiotic stresses, but their roles in mediating elicitor‐induced disease resistance remains largely unexplored. PevD1 is a Verticillium dahliae secretory effector that can induce disease resistance in cotton and tobacco plants. In our previous work, Nicotiana benthamiana ERF114 ( NbERF114) was identified in a screen of genes differentially expressed in response to PevD1 infiltration. Here, we found that the ortholog of NbERF114 in Arabidopsis thaliana ( ERF114) also strongly responded to PevD1 treatment and transcripts were induced by Pseudomonas syringae pv. tomato (Pst) DC3000 infection. Loss of ERF114 function caused impaired disease resistance, while overexpressing ERF114 ( OE‐ERF114) enhanced resistance to Pst DC3000. Moreover, ERF114 mediated PevD1‐induced disease resistance. RNA‐sequencing analysis revealed that the transcript level of phenylalanine ammonia‐lyase1 ( PAL1) and its downstream genes were significantly suppressed in erf114 mutants compared with A. thaliana Col‐0. Reverse transcription‐quantitative PCR (RT‐qPCR) analysis further confirmed that the PAL1 mRNA level was significantly elevated in overexpressing OE‐ERF114 plants but reduced in erf114 mutants compared with Col‐0. Chromatin immunoprecipitation‐qPCR (ChIP‐qPCR) and electrophoretic mobility shift assay verified that ERF114 directly bound to the promoter of PAL1. The gene expression profiles of ERF114 and PAL1 in oestradiol‐inducible transgenic plants confirmed ERF114 could activate PAL1 transcriptional expression. Further investigation revealed that ERF114 positively modulated PevD1‐induced lignin and salicylic acid accumulation, probably by activating PAL1 transcription.
Abstract
Related collections
Most cited references58

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data
- Record: found
- Abstract: found
- Article: not found
The plant immune system.
- Record: found
- Abstract: found
- Article: not found
