- Record: found
- Abstract: found
- Article: found
Plant compartment and genetic variation drive microbiome composition in switchgrass roots
Read this article at
Summary
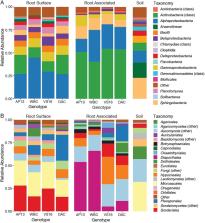
Switchgrass ( Panicum virgatum) is a promising biofuel crop native to the United States with genotypes that are adapted to a wide range of distinct ecosystems. Various plants have been shown to undergo symbioses with plant growth‐promoting bacteria and fungi, however, plant‐associated microbial communities of switchgrass have not been extensively studied to date. We present 16S ribosomal RNA gene and internal transcribed spacer (ITS) data of rhizosphere and root endosphere compartments of four switchgrass genotypes to test the hypothesis that host selection of its root microbiota prevails after transfer to non‐native soil. We show that differences in bacterial, archaeal and fungal community composition and diversity are strongly driven by plant compartment and switchgrass genotypes and ecotypes. Plant‐associated microbiota show an enrichment in Alphaproteobacteria and Actinobacteria as well as Sordariales and Pleosporales compared with the surrounding soil. Root associated compartments display low‐complexity communities dominated and enriched in Actinobacteria, in particular Streptomyces, in the lowland genotypes, and in Alphaproteobacteria, specifically Sphingobium, in the upland genotypes. Our comprehensive root analysis serves as a snapshot of host‐specific bacterial and fungal associations of switchgrass in the field and confirms that host‐selected microbiomes persist after transfer to non‐native soil.
Related collections
Most cited references45

- Record: found
- Abstract: found
- Article: found
FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments
- Record: found
- Abstract: found
- Article: not found
Integration of biological networks and gene expression data using Cytoscape.
- Record: found
- Abstract: found
- Article: not found