- Record: found
- Abstract: found
- Article: found
The role of biogeographical barriers on the historical dynamics of passerine birds with a circum‐Amazonian distribution

Read this article at
Abstract
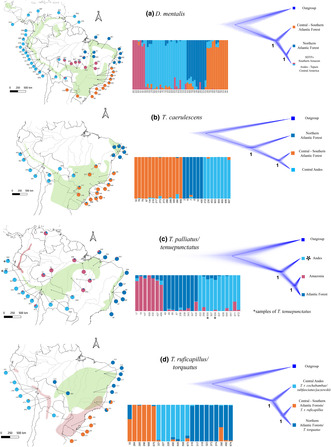
Common distributional patterns have provided the foundations of our knowledge of Neotropical biogeography. A distinctive pattern is the “circum‐Amazonian distribution”, which surrounds Amazonia across the forested lowlands south and east of the basin, the Andean foothills, the Venezuelan Coastal Range, and the Tepuis. The underlying evolutionary and biogeographical mechanisms responsible for this widespread pattern of avian distribution have yet to be elucidated. Here, we test the effects of biogeographical barriers in four species in the passerine family Thamnophilidae by performing comparative demographic analyses of genome‐scale data. Specifically, we used flanking regions of ultraconserved regions to estimate population historical parameters and genealogical trees and tested demographic models reflecting contrasting biogeographical scenarios explaining the circum‐Amazonian distribution. We found that taxa with circum‐Amazonian distribution have at least two main phylogeographical clusters: (1) Andes, often extending into Central America and the Tepuis; and (2) the remaining of their distribution. These clusters are connected through corridors along the Chaco–Cerrado and southeastern Amazonia, allowing gene flow between Andean and eastern South American populations. Demographic histories are consistent with Pleistocene climatic fluctuations having a strong influence on the diversification history of circum‐Amazonian taxa, Refugia played a crucial role, enabling both phenotypic and genetic differentiation, yet maintaining substantial interconnectedness to keep considerable levels of gene flow during different dry/cool and warm/humid periods. Additionally, steep environmental gradients appear to play a critical role in maintaining both genetic and phenotypic structure.
Abstract
This study found similarities at populational, phylogenetic, and evolutionary levels among four taxonomic groups of passerine birds with a circum‐Amazonian distribution. The presence of Refugia result of climatic oscillations during the Pleistocene was the main drive in the diversification of these circum‐Amazonian taxa.
Related collections
Most cited references177

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data

- Record: found
- Abstract: found
- Article: found
The Sequence Alignment/Map format and SAMtools

- Record: found
- Abstract: found
- Article: found