- Record: found
- Abstract: found
- Article: found
The Gut Microbiota Protects Bees from Invasion by a Bacterial Pathogen
Read this article at
ABSTRACT
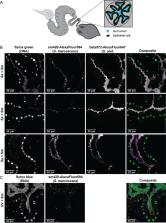
Commensal microbes in animal guts often help to exclude bacterial pathogens. In honey bees, perturbing or depleting the gut microbiota increases host mortality rates upon challenge with the opportunistic pathogen Serratia marcescens, suggesting antagonism between S. marcescens and one or more members of the bee gut microbiota. In laboratory culture, S. marcescens uses a type VI secretion system (T6SS) to kill bacterial competitors, but the role of this T6SS within hosts is unknown. Using infection assays, we determined how the microbiota impacts the abundance and persistence of S. marcescens in the gut and visualized colocalization of S. marcescens with specific community members in situ. Using T6SS-deficient S. marcescens strains, we measured T6SS-dependent killing of gut isolates in vitro and compared the persistence of mutant and wild-type strains in the gut. We found that S. marcescens is rapidly eliminated in the presence of the microbiota but persists in microbiota-free guts. Protection is reduced in monocolonized and antibiotic-treated bees, possibly because different symbionts occupy distinct niches. Serratia marcescens uses a T6SS to antagonize Escherichia coli and other S. marcescens strains but shows limited ability to kill bee symbionts. Furthermore, wild-type and T6SS-deficient S. marcescens strains achieved similar abundance and persistence in bee guts. Thus, an intact gut microbiota offers robust protection against this common pathogen, whose T6SSs do not confer the ability to compete with commensal species.
IMPORTANCE Bacteria living within guts of animals can provide protection against infection by pathogens. Some pathogens have been shown to use a molecular weapon known as a T6SS to kill beneficial bacteria during invasion of the mouse gut. In this study, we examined how bacteria native to the honey bee gut work together to exclude the opportunistic pathogen Serratia marcescens. Although S. marcescens has a T6SS that can kill bacteria, bee gut bacteria seem resistant to its effects. This limitation may partially explain why ingestion of S. marcescens is rarely lethal to insects with healthy gut communities.
Related collections
Most cited references60
- Record: found
- Abstract: found
- Article: not found
Fiji: an open-source platform for biological-image analysis.
- Record: found
- Abstract: found
- Article: found
ImageJ2: ImageJ for the next generation of scientific image data

- Record: found
- Abstract: found
- Article: found