- Record: found
- Abstract: found
- Article: found
Foot-and-mouth disease virus VP1 degrades YTHDF2 through autophagy to regulate IRF3 activity for viral replication
Read this article at
ABSTRACT
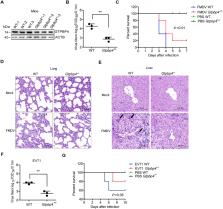
Many viruses, including foot-and-mouth disease virus (FMDV), can promote the degradation of host proteins through macroautophagy/autophagy, thereby promoting viral replication. However, the regulatory mechanism between autophagy and innate immune responses is not fully understood during FMDV infection. Here, we found that the host GTPBP4/NOG1 (GTP binding protein 4) is a negative regulator of innate immune responses. GTPBP4 deficiency promotes the antiviral innate immune response, resulting in the ability of GTPBP4 to promote FMDV replication. Meanwhile, GTPBP4-deficient mice are more resistant to FMDV infection. To antagonize the host’s antiviral immunity, FMDV structural protein VP1 promotes the expression of GTPBP4, and the 209th site of VP1 is responsible for this effect. Mechanically, FMDV VP1 promotes autophagy during virus infection and interacts with and degrades YTHDF2 (YTH N6-methyladenosine RNA binding protein F2) in an AKT-MTOR-dependent autophagy pathway, resulting in an increase in GTPBP4 mRNA and protein levels. Increased GTPBP4 inhibits IRF3 binding to the Ifnb/Ifn-β promoter, suppressing FMDV-induced type I interferon production. In conclusion, our study revealed an underlying mechanism of how VP1 negatively regulates innate immunity through the autophagy pathway, which would contribute to understanding the negative regulation of host innate immune responses and the function of GTPBP4 and YTHDF2 during FMDV infection.
Abbreviation: 3-MA:3-methyladenine; ACTB: actin beta; ATG: autophagy related; ChIP:chromatin immunoprecipitation; CQ: chloroquine; DAPI:4’,6-diamidino-2-phenylindole; dpi: days post-infection; EV71:enterovirus 71; FMDV: foot-and-mouth disease virus; GTPBP4/NOG1: GTPbinding protein 4; HIF1A: hypoxia inducible factor 1 subunit alpha;hpt:hours post-transfection; IFNB/IFN-β:interferon beta; IRF3: interferon regulatory factor 3; MAP1LC3/LC3:microtubule associated protein 1 light chain 3; MAVS: mitochondriaantiviral signaling protein; MOI: multiplicity of infection; MTOR:mechanistic target of rapamycin kinase; m6A: N(6)-methyladenosine;qPCR:quantitativePCR; SIRT3:sirtuin 3; SQSTM1/p62: sequestosome 1; STING1: stimulator ofinterferon response cGAMP interactor 1; siRNA: small interfering RNA;TBK1: TANK binding kinase 1; TCID 50:50% tissue culture infectious doses; ULK1: unc-51 like autophagyactivating kinase 1; UTR: untranslated region; WT: wild type; YTHDF2:YTH N6-methyladenosine RNA binding protein F2
Related collections
Most cited references92
- Record: found
- Abstract: found
- Article: not found
Analyzing real-time PCR data by the comparative C(T) method.
- Record: found
- Abstract: found
- Article: not found
Autophagy: cellular and molecular mechanisms.
- Record: found
- Abstract: found
- Article: found