- Record: found
- Abstract: found
- Article: found
C9orf72 hexanucleotide repeat allele tagging SNPs: Associations with ALS risk and longevity
Read this article at
Abstract
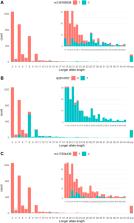
C9orf72 hexanucleotide repeat expansion is a common cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). The C9orf72 locus may harbor residual risk outside the hexanucleotide repeat expansion, but the evidence is conflicting. Here, we first compared 683 unrelated amyotrophic lateral sclerosis cases and 3,196 controls with Finnish ancestry to find best single nucleotide polymorphisms that tag the C9orf72 hexanucleotide repeat expansion and intermediate-length alleles. Rs2814707 was the best tagging single nucleotide polymorphisms for intermediate-length alleles with ≥7 repeats ( p = 5 × 10 −307) and rs139185008 for the hexanucleotide repeat expansion ( p = 7 × 10 −114) as well as alleles with ≥20 repeats. rs139185008*C associated with amyotrophic lateral sclerosis after removing cases with the hexanucleotide repeat expansion, especially in the subpopulation homozygous for the rs2814707*T ( p = 0.0002, OR = 5.06), which supports the concept of residual amyotrophic lateral sclerosis risk at the C9orf72 haplotypes other than the hexanucleotide repeat expansion. We then leveraged Finnish biobank data to test the effects of rs2814707*T and rs139185008*C on longevity after removing individuals with amyotrophic lateral sclerosis / frontotemporal dementia diagnoses. In the discovery cohort ( n = 230,006), the frequency of rs139185008*C heterozygotes decreased significantly with age in the comparisons between 50 and 80 years vs. >80 years ( p = 0.0005) and <50 years vs. >80 years ( p = 0.0001). The findings were similar but less significant in a smaller replication cohort (2-sided p = 0.037 in 50–80 years vs. >80 years and 0.061 in <50 years vs. >80 years). Analysis of the allele frequencies in 5-year bins demonstrated that the decrease of rs139185008*C started after the age of 70 years. The hexanucleotide repeat expansion tagging single nucleotide polymorphisms decreasing frequency with age suggests its’ association with age-related diseases probably also outside amyotrophic lateral sclerosis / frontotemporal dementia.
Related collections
Most cited references31

- Record: found
- Abstract: found
- Article: found
Second-generation PLINK: rising to the challenge of larger and richer datasets
- Record: found
- Abstract: found
- Article: found
A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD.

- Record: found
- Abstract: found
- Article: found