- Record: found
- Abstract: found
- Article: not found
Fast Bayesian parameter estimation for stochastic logistic growth models

Read this article at
Abstract
Abstract
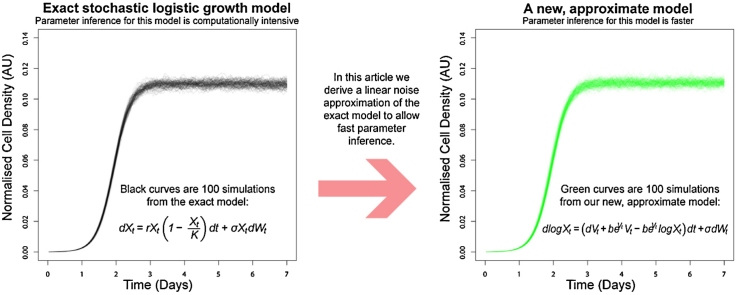
The transition density of a stochastic, logistic population growth model with multiplicative intrinsic noise is analytically intractable. Inferring model parameter values by fitting such stochastic differential equation (SDE) models to data therefore requires relatively slow numerical simulation. Where such simulation is prohibitively slow, an alternative is to use model approximations which do have an analytically tractable transition density, enabling fast inference. We introduce two such approximations, with either multiplicative or additive intrinsic noise, each derived from the linear noise approximation (LNA) of a logistic growth SDE. After Bayesian inference we find that our fast LNA models, using Kalman filter recursion for computation of marginal likelihoods, give similar posterior distributions to slow, arbitrarily exact models. We also demonstrate that simulations from our LNA models better describe the characteristics of the stochastic logistic growth models than a related approach. Finally, we demonstrate that our LNA model with additive intrinsic noise and measurement error best describes an example set of longitudinal observations of microbial population size taken from a typical, genome-wide screening experiment.
Related collections
Most cited references24
- Record: found
- Abstract: found
- Article: not found
Stochastic modelling for quantitative description of heterogeneous biological systems.
- Record: found
- Abstract: found
- Article: not found
Bayesian inference for stochastic kinetic models using a diffusion approximation.
- Record: found
- Abstract: not found
- Article: not found