- Record: found
- Abstract: found
- Article: found
Increased incompatibility of heterologous algal symbionts under thermal stress in the cnidarian-dinoflagellate model Aiptasia
Read this article at
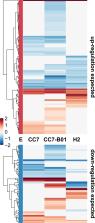
Abstract
Rising ocean temperatures are increasing the rate and intensity of coral mass bleaching events, leading to the collapse of coral reef ecosystems. To better understand the dynamics of coral-algae symbioses, it is critical to decipher the role each partner plays in the holobiont’s thermotolerance. Here, we investigated the role of the symbiont by comparing transcriptional heat stress responses of anemones from two thermally distinct locations, Florida (CC7) and Hawaii (H2) as well as a heterologous host-symbiont combination composed of CC7 host anemones inoculated with the symbiont Breviolum minutum (SSB01) from H2 anemones (CC7-B01). We find that oxidative stress and apoptosis responses are strongly influenced by symbiont type, as further confirmed by caspase-3 activation assays, but that the overall response to heat stress is dictated by the compatibility of both partners. Expression of genes essential to symbiosis revealed a shift from a nitrogen- to a carbon-limited state only in the heterologous combination CC7-B01, suggesting a bioenergetic disruption of symbiosis during stress. Our results indicate that symbiosis is highly fine-tuned towards particular partner combinations and that heterologous host-symbiont combinations are metabolically less compatible under stress. These results are essential for future strategies aiming at increasing coral resilience using heterologous thermotolerant symbionts.
Abstract
Algal symbionts are demonstrated to improve heat-stress tolerance in anemone hosts only when the two partners are closely compatible.
Related collections
Most cited references68
- Record: found
- Abstract: found
- Article: not found
Near-optimal probabilistic RNA-seq quantification.
- Record: found
- Abstract: found
- Article: not found
Apoptosis: a review of programmed cell death.
- Record: found
- Abstract: found
- Article: not found